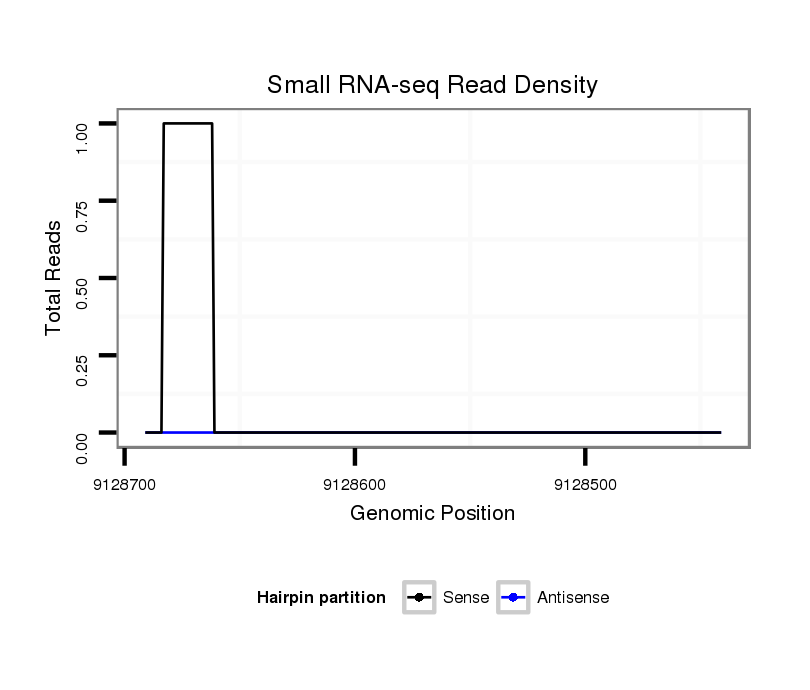
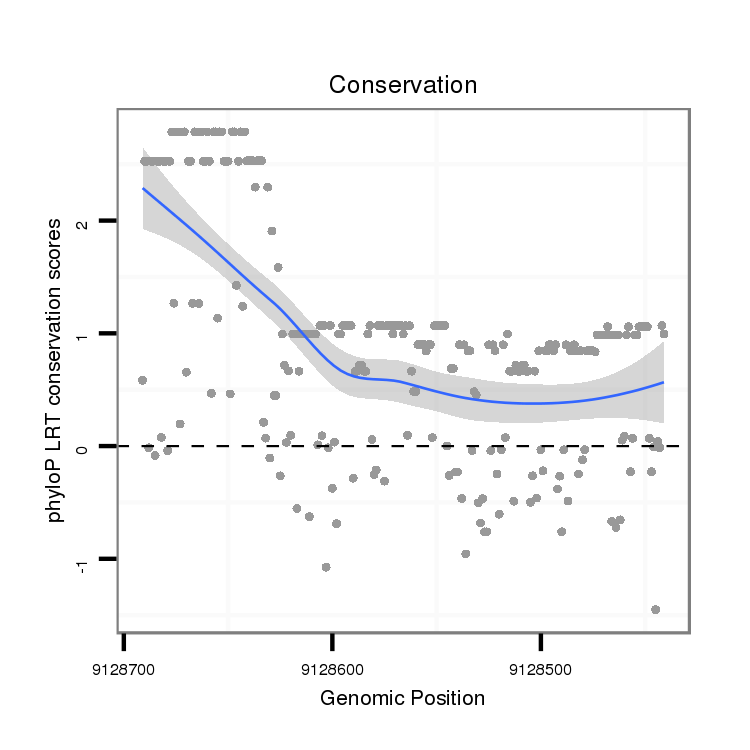
| CGCCGCGCCGGCCGAAAATTTCCCCTTCTGCACCATTAAGCCCAATGAGAATAAGTATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGGCC-----------------------ATTGGA----GTCGCAATTTGCGTTGCCTCC--A---AATAATTGTAATCAATG------------CAATTTTT----------TGTCCGTTCGGGGCATTGCATT-------------------------GCCGCAGCTTCCCTGTGTCGCTTCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGGCGTG----TCCCAATCT---------CGGCAATT-TGCCAGATC | Size | Hit Count | Total Norm | Total | M046
Female-body | V041
Embryo | V049
Head | V056
Head | M060
Embryo | V110
Male-body |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGG................................................................................................................................................................................................................................................................................................................................ | 25 | 1 | 49.00 | 49 | 48 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................................................................................................................................................................................TTCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGG............................................. | 25 | 1 | 23.00 | 23 | 22 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................................................................................................................................................................................TTCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GG.............................................. | 24 | 1 | 23.00 | 23 | 18 | 0 | 0 | 0 | 4 | 1 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGGC............................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 22.00 | 22 | 20 | 0 | 0 | 0 | 2 | 0 |
| ...............................................................................................................................................................................................................................................TGTAATCAATG------------CAATTTTT----------TGTCCGT............................................................................................................................................................................................................ | 26 | 1 | 21.00 | 21 | 20 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................................................................................................................................................................................................TCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGG............................................. | 24 | 1 | 14.00 | 14 | 12 | 1 | 0 | 0 | 0 | 1 |
| ..........................TCTGCACCATTAAGCCCAATGAGAG........................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 1 | 10.00 | 10 | 6 | 0 | 0 | 0 | 4 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................TCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GG.............................................. | 23 | 1 | 9.00 | 9 | 8 | 0 | 0 | 0 | 1 | 0 |
| ..........................TCTGCACCATTAAGCCCAATGAGAAT....................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 9.00 | 9 | 7 | 0 | 0 | 0 | 2 | 0 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCG................................................................................................................................................................................................................................................................................................................................. | 24 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................TTCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------G............................................... | 23 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCC.................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 7.00 | 7 | 2 | 0 | 0 | 0 | 5 | 0 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGGCC-----------------------A...................................................................................................................................................................................................................................................................................................... | 28 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGGCC-----------------------AT..................................................................................................................................................................................................................................................................................................... | 29 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGG................................................................................................................................................................................................................................................................................................................................ | 24 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................TCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------G............................................... | 22 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 | 0 | 0 |
| ..........................TCTGCACCATTAAGCCCAATGAGAGC....................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 5.00 | 5 | 0 | 0 | 0 | 0 | 5 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................TCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGGC........................................... | 26 | 1 | 5.00 | 5 | 3 | 0 | 0 | 0 | 2 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................TTCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGG............................................ | 26 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| .......................................................TATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATG....................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4 | 3 | 0 | 0 | 0 | 0 | 1 |
| .........................................................TTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGC................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ..........................TCTGCACCATTAAGCCCAATGAGAATA...................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................TCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGG............................................ | 25 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ....................TCCCCTTCTGCACCATTAAGCCCAATGA........................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..........................TCTGCACCATTAAGCCCAATGAGA......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................TCCCTGTGTCGCTTCTAT---------GTTGGC.......................................................................................................................... | 24 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCC.................................................................................................................................................................................................................................................................................................................................. | 29 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 1 | 0 |
| .................................................................TACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGGC............................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ....................TCCCCTTCTGCACCATTAAGCCCAATGAG.......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATG....................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................TGTAATCAATG------------CAATTTTT----------TGTCCG............................................................................................................................................................................................................. | 25 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAAGTATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGC.............................................................................................................................................................................................................................................................................................................................................. | 24 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................TGTAATCAATG------------CAATTTTT----------TGTCCGTC........................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 1 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGC................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................CTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GG.............................................. | 22 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCC.................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................TCCCCTTCTGCACCATTAAGCCCA............................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................GTAATCAATG------------CAATTTTT----------TGTCCGT............................................................................................................................................................................................................ | 25 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGA................................................................................................................................................................................................................................................................................................................................ | 25 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAAGTATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGA......................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAAGTATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCT............................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................TCTGCACCATTAAGCCCAATGAGAA........................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................TCCCTGTGTCGCTTCTAT---------GTTGG........................................................................................................................... | 23 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................TTTCCCCTTCTGCACCATTAAGCCCA............................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAAGTATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTG............................................................................................................................................................................................................................................................................................................................................ | 25 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................TCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGGCG.......................................... | 27 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................TTCCCCTTCTGCACCATTAAGCCCAATG............................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 28 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................CTGCACCATTAAGCCCAATGAGAAT....................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCG.................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................TCTGCACCATTAAGCCCAATGAG.......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TTGCATT-------------------------GCCGCAGCTTCCCTC..................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGGCC-----------------------AT..................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................TGTAATCAATG------------CAATTTTT----------TGTCCGTTC.......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGC................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................CTGCACCATTAAGCCCAATGAGAG........................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGGCC-----------------------T...................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................TCGGGGCATTGCATT-------------------------GCCGCAGCTTC......................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................TC-------------GCG---------------------------------------------------GGGGCGTG----TCCCAATCT---------CGGC.............. | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................AATCAATG------------CAATTTTT----------TGTCCGTC........................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......GCCGGCCGAAAATTTCCCCTTCTGCACCA........................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................TCCCCTTCTGCACCATTAAGCCC................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................A-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGG................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................TGCACCATTAAGCCCAATGAGAG........................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................T---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGGCGTG----T................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................TG----TCCCAATCT---------CGGCAATT-TGCCAGATC | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGGCC-----------------------....................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................TGTCCGTTCGGGGCATTGCATT-------------------------GCCGCA.............................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCC..................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................TGTCCGTTCGGGGCATTGCATT-------------------------G................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................TATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGC...................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................T---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGGCGTG----.................................... | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................TAATCAATG------------CAATTTTT----------TGTCCGTTCGG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................TCAATG------------CAATTTTT----------TGTCCGTTCGGGGCA.................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................TAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGG............................................. | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................TCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGGG........................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................TAAGTATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCT............................................................................................................................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCG................................................................................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................TTCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGGC........................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................TC-------------GCG---------------------------------------------------GGGGCGTG----TCCCAATCT---------CG................ | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................TTCTAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------................................................ | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................GTAATCAATG------------CAATTTTT----------TGTCCG............................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................A-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCC.................................................................................................................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGAT........................................................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................TAT---------GTTGGCTGCCGTC-------------GCG---------------------------------------------------GGGGC........................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................ACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGG................................................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................A---------------------------------------------------------------------------------AG-AGCTGCGATGCCGCCGG................................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................TTGGA----GTCGCAATTTGCGTC.............................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................TCCCTGTGTCGCTTCTAT---------GTTGGCTG........................................................................................................................ | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGC...................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................TTCCCTGTGTCGCTTCTAT---------GTTGGCTG........................................................................................................................ | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCCT.................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................CATTAAGCCCAATGAGAAT....................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................TTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGATGCC..................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AAGTATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCT............................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAAGTATTCCAA-TTACCA---------------------------------------------------------------------------------AG-AGCTGCGA......................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TCGCAATTTGCGTTGCCTCC--A---AATA.............................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................TGTAATCAATG------------CAATTTTT----------TGTCCGTT........................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................TCCCCTTCTGCACCATTAAGCCCAAT............................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................TTCTGCACCATTAAGCCCAATGAG.......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................ATTAAGCCCAATGAGAAA....................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |