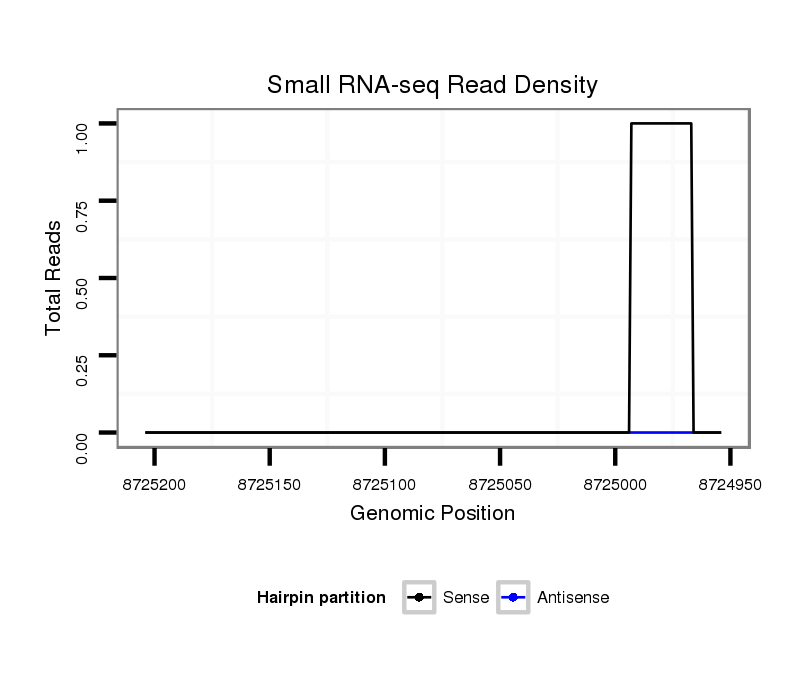
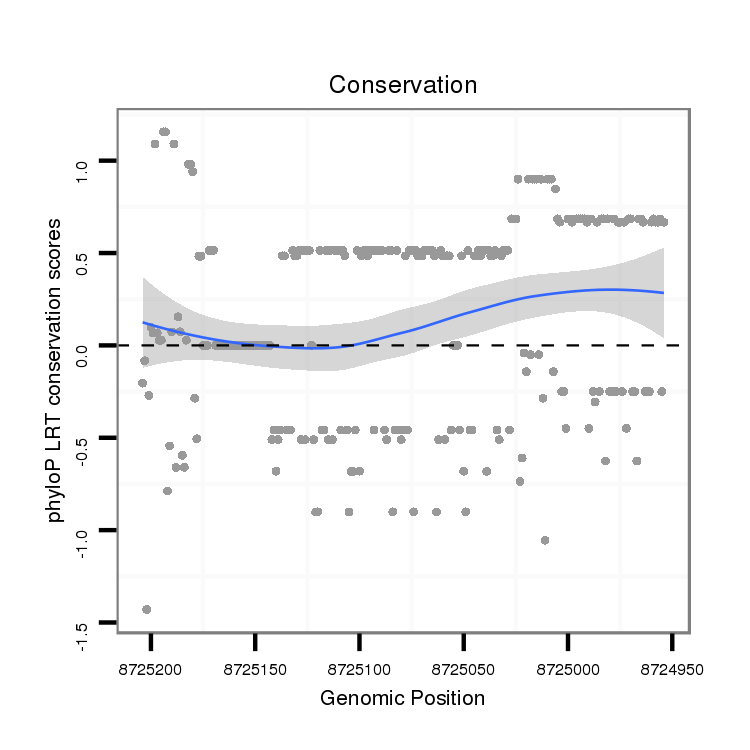
ID:dvi_13679 |
Coordinate:scaffold_12970:8725004-8725154 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ18734-cds]; exon [dvir_GLEANR_3560:6]; intron [Dvir\GJ18734-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTTTCCCTTTTTCCTGTTAGTAAACTGTAGTTTTATGATATTGTGTTCATGGGGGCCTTTTTTGGTCCCAGCAGCATTTAAAACGTGTATTATTTGAGAGCTGAGCTTCTAGTTTTGATAGGATAAGGTTGAGGGTATATCCTAGACGGTAGACGGATCCGACTAAAGCATACTTATAATTATTTTTATTATCCTTTACAGAAAAAGAAAATTCAAATGGAACAAGAAGCAGAAAATGTTGGGTCACGTGC **************************************************.((((((......))))))..((((.((((((..........)))))).))))................((((((((((((.((((((....(((.(((.........))).)))....)))))).))))).......)))))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060662 9x160_0-2h_embryos_total |
GSM1528803 follicle cells |
SRR060684 140x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
V116 male body |
M028 head |
M027 male body |
SRR060681 Argx9_testes_total |
SRR060659 Argentina_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060688 160_ovaries_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................TTCAAATGGAACAAGAAGCAGAAAATG............. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AGTGTATATCCTAGAC........................................................................................................ | 16 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................GATATTACGTTGATGGGGGCC.................................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................ATATTACGTTGATGGGGGCC.................................................................................................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AGTGTATATCCTAGACC....................................................................................................... | 17 | 2 | 18 | 0.22 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................TAGACGGATACGAATAAGGC.................................................................................. | 20 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................CGGTAGACGTAGTCGACT....................................................................................... | 18 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................AGAAGCACAAAATATCGGGT....... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................ATCGGATATGGTTGAGGGT................................................................................................................... | 19 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ATATGGTTGAGGGTATAG............................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AGATGGATCCGAATAAGGC.................................................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TGCTTTGATAGGGTAAGGT.......................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................GATAGAATAAAGTTGAGGGA................................................................................................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................TGTGTTCATGGTGGGCTCT............................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................TTCAGATGGAACAAAAAG...................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................TCGGTAGAGGGATACGAC........................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TTTGGTTTCAGCAGCATT............................................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
AAAAGGGAAAAAGGACAATCATTTGACATCAAAATACTATAACACAAGTACCCCCGGAAAAAACCAGGGTCGTCGTAAATTTTGCACATAATAAACTCTCGACTCGAAGATCAAAACTATCCTATTCCAACTCCCATATAGGATCTGCCATCTGCCTAGGCTGATTTCGTATGAATATTAATAAAAATAATAGGAAATGTCTTTTTCTTTTAAGTTTACCTTGTTCTTCGTCTTTTACAACCCAGTGCACG
**************************************************.((((((......))))))..((((.((((((..........)))))).))))................((((((((((((.((((((....(((.(((.........))).)))....)))))).))))).......)))))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
SRR1106718 embryo_6-8h |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................GAAATAACCAGGATCTTCGT............................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CAGGAAAGGTCTTCTTCTTT......................................... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................AAAAATCAGGGTCGTC................................................................................................................................................................................. | 16 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................AAAAAACCAGGGTTGTAG................................................................................................................................................................................ | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................AAAATAATAGGCAAGGTC.................................................. | 18 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CCATGTGCCTAGGCTAGT...................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:8724954-8725204 - | dvi_13679 | TTTTCCCT-------TTTTCCTGTTAGTAAACTGTAGTTTTATGATATTGTGTTCATGGGGGCCTTTTTTGGTCCCAGCAGCATTTAAAACGTGTATTATTTGAGAGCTGAGCTTCTAGTTTTGATA--GGATAAGGTTGAGGGTATATCCTAGACGGTAGACGGATCCGACTAAAGCATACTTATAATTATTTTTATTATCCTTTACAGAAAAAGAA---------AATTCAAATGGA------ACAAGAAGCAGAAAATGTTGGGTC---ACGTGC |
| droMoj3 | scaffold_6482:276766-276801 - | CTCATCCCGATCTTTTTTTATCGATAGAAAACCTTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15252:5195459-5195622 + | CTGTCTCTGTTTCTCAATTATCGATACAT----------TTA---------------------------AATATCCGATAGCAATAAA-TGCTACAATTTTTAAGGCAGAATCTTCTAATTTTATTAATCAACTGGATTCAGGGTATATCGAAGTCGA---GCTCACTCGACTACAGCACTCTTAC---TTGTTTTATTATTGTTTAC---------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004909:10304136-10304231 - | TCAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATTGTCGTAATTTTTTTTTCCAGGGCAAGAAAGGTGAGGAAATTCCAGAGAAGGCTCGAGAGAAGGCGGCAAGCCTTGAACCTCAACGTAC | |
| droEre2 | scaffold_4845:16850148-16850174 + | CACGCCCT-------TTTTTACGCCCACAAACCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:49 PM