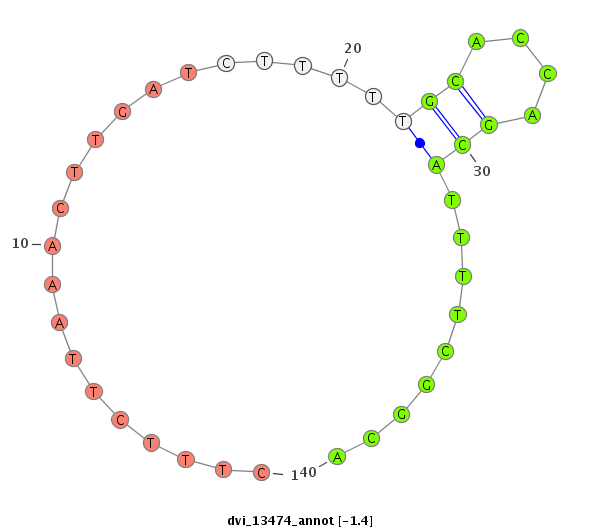
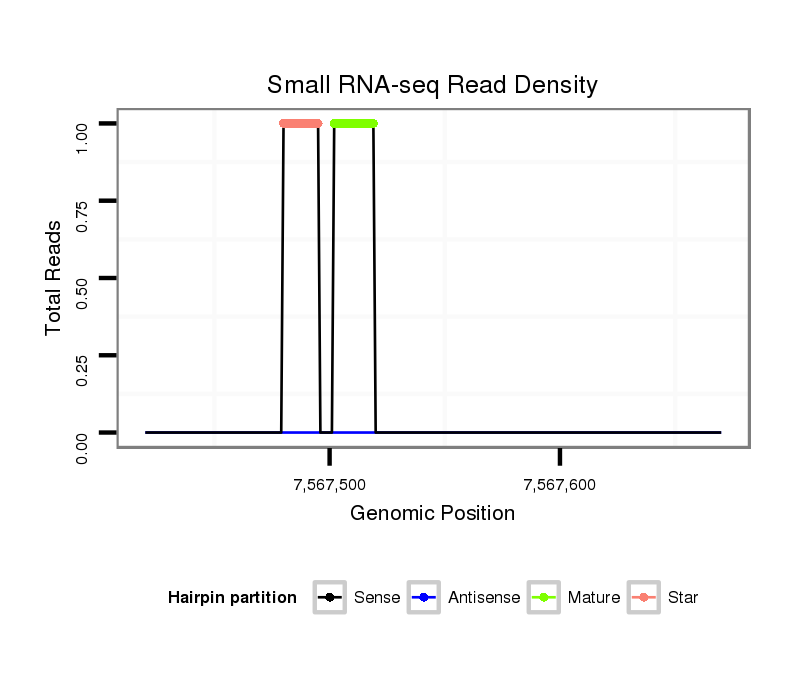

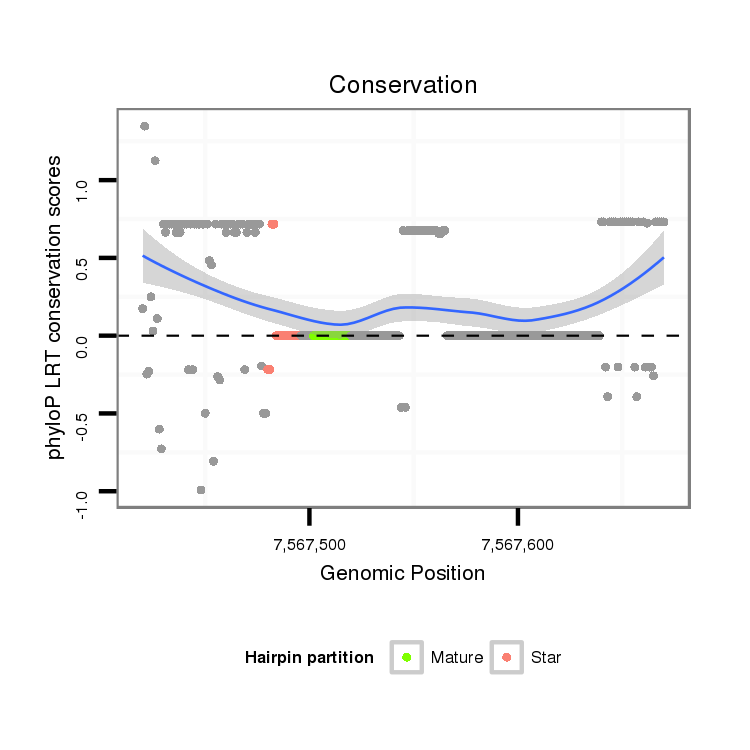
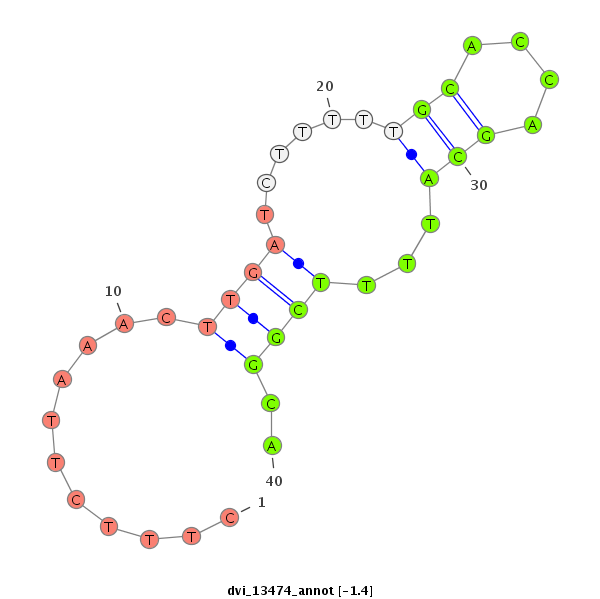
ID:dvi_13474 |
Coordinate:scaffold_12970:7567470-7567620 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -1.4 | -1.0 | -1.0 |
 |
 |
 |
CDS [Dvir\GJ19360-cds]; exon [dvir_GLEANR_4250:1]; intron [Dvir\GJ19360-in]
| Name | Class | Family | Strand |
| (T)n | Simple_repeat | Simple_repeat | + |
| --------------------------------------############--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATCTTTACCCAGTAATCTCATTAAAAAAAAAAACGAATATGTATCCATACGTAAGTAGAACTTTCTTAAACTTGATCTTTTTGCACCAGCATTTTCGGCAATAGATGCACTTATATATTTTTTTTTTTTTTTTTTTTTTTTTGGTATCACAATTTTCATTTAAATAGTTGTCGAATGTACGCTTCTTAGATACATGTCGCTCGTTTTTTTTTTTTTGTCTTTTCTTTTCATAAAATACAAACCATTTTTTT ************************************************************.....................(((....))).........******************************************************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060667 160_females_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR060669 160x9_females_carcasses_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................GCACCAGCATTTTCGGCA....................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................CTTTCTTAAACTTGAT............................................................................................................................................................................... | 16 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................TTTGTCTATTCTTTTTATAAA................. | 21 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................GTCGCACGTACGGTTCTTA............................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................TTGTTTTTTTTTGTCTTTTC........................... | 20 | 1 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................TAAAAAAAAAATTGAATATGTA................................................................................................................................................................................................................ | 22 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TAGAAATGGGTCATTAGAGTAATTTTTTTTTTTGCTTATACATAGGTATGCATTCATCTTGAAAGAATTTGAACTAGAAAAACGTGGTCGTAAAAGCCGTTATCTACGTGAATATATAAAAAAAAAAAAAAAAAAAAAAAAACCATAGTGTTAAAAGTAAATTTATCAACAGCTTACATGCGAAGAATCTATGTACAGCGAGCAAAAAAAAAAAAACAGAAAAGAAAAGTATTTTATGTTTGGTAAAAAAA
*******************************************************************************************************************************************************.....................(((....))).........************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060674 9x140_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
M047 female body |
SRR060665 9_females_carcasses_total |
M061 embryo |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................AAAAAAAAAACCATTGTGTTAAAAA.............................................................................................. | 25 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............AAAGTAATTTTTTTTTTTGCTCATACGT................................................................................................................................................................................................................ | 28 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................ACGTGGGCGTCAAAGCCATTA..................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AAAAAAAAAAACCATAGGGTTGACA............................................................................................... | 25 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGGGCCATTAGGGTAATTT.................................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AAAAAAAAAAAAACCATAGGGTTGA................................................................................................. | 25 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAAAAAAACCATAGGGTTGA................................................................................................. | 21 | 2 | 5 | 0.40 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGGGCCATTAGGGTAATT................................................................................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................AAAAAAAAAAAACCATAGGGTTG.................................................................................................. | 23 | 2 | 7 | 0.29 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................AAAAAAAACCATAGGGTTGA................................................................................................. | 20 | 2 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGGGCCATTAGGGTAATTTTA................................................................................................................................................................................................................................ | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AAAACATAAAACCATAGAGTTAA................................................................................................. | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AAAAAAAAAACCATAGGGTTGA................................................................................................. | 22 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................ACGTGGGCGTCAAAGCCAT....................................................................................................................................................... | 19 | 3 | 9 | 0.22 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AAAAAAAAAACCATAGGGTTGACA............................................................................................... | 24 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AAAAAAAAAAGCCCGAAAAGAAAAGT..................... | 26 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................AAAAAACCATAGGGTTGA................................................................................................. | 18 | 2 | 17 | 0.12 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................AAAAAAAAAAAAAAAAAACCATA........................................................................................................ | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ......................................................................................................................................AAAAAAAACCATAGGGTTGACA............................................................................................... | 22 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................AAAAAAAAAAAAAAAAAAAACCATA........................................................................................................ | 25 | 0 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................AAAAAAAAAAAAAAAAAAAACCA.......................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................TGAAAGAGAAAAACGTGG.................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................................TATGTCCAGCGAGTAAAAG........................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................TCATCACCATACATGCGA.................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:7567420-7567670 + | dvi_13474 | ATCTTTACCCAGTAATCTCATTAAAAA-AAAA----AACGAATATGTATCCATACGTAAGTAGAACTTTCTTAAACTTGATCTTTTTGCACCAGCATTTTCGGCAATAGATGCACTTATATATTTTTTTTTTTTTTTTTTTTTTTTTGGTATCACAATTTTCATTTAAATAGTTGTCGAATGTACGCTTCTTAGATACATGTCGCTCGTTTTTTTTTTTTTGTCTTTTCTTTTCATAAAATACAAACCATTTTTTT |
| droMoj3 | scaffold_6482:2153877-2153945 - | ATTCTTACAGAGTAATCTCATTAAAAACATACAAATAACCATCATGTATCCATATGTAAGTAGCCTTTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15203:7044746-7044812 - | ATCCTAACCCAGTAATCTCATTTATAAAACAAAACTA--CAACATGTATCCATACGTAAGTAAAACATT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_48:237294-237320 - | TTTTT----------------------------------------------------------------------------------------------------------------------------GTGTTTTTTTTTTTTTTTGGTA--------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000777354:2265-2305 + | TTCTATATAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCATTTTTATAAAATGAAAATCGCATTTTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/17/2015 at 04:45 AM