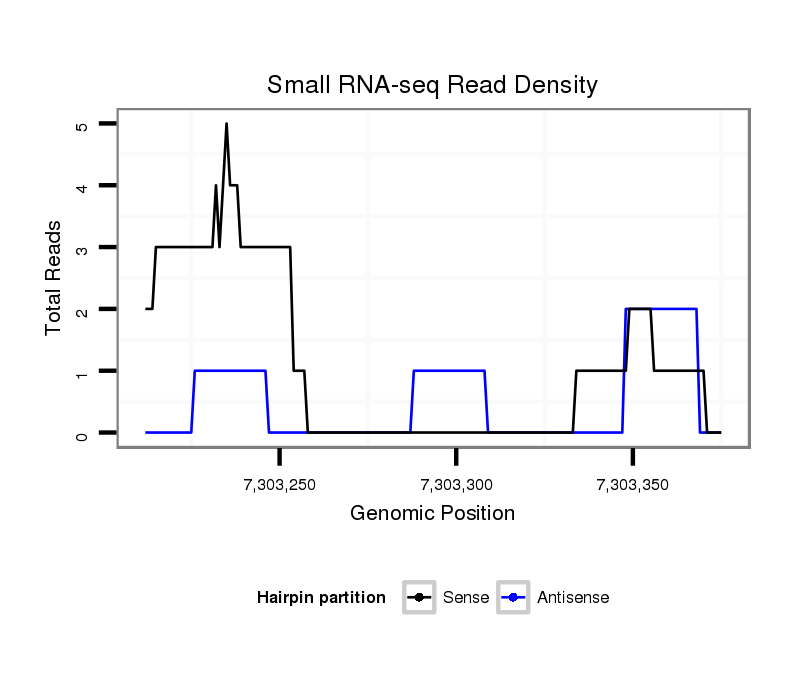
ID:dvi_13426 |
Coordinate:scaffold_12970:7303262-7303325 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_4237:3]; CDS [Dvir\GJ19346-cds]; exon [dvir_GLEANR_4237:2]; CDS [Dvir\GJ19346-cds]; intron [Dvir\GJ19346-in]
No Repeatable elements found
| ##################################################----------------------------------------------------------------################################################## AATCAACCACACGATGCAGAAAGTACAGCTGGATGCAAATGTGCCTGAAGGTATGTTACATGACAAACAGCTTTTAATTTCCCTGCTAAATCTCCTTCTCAATCTGAATTACAGAAACTCCAGAGAAGACAACAGCGACGTCAAACAAGCAGCGCAACAATCGC **************************************************....................((...(((((.........................)))))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060665 9_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060675 140x9_ovaries_total |
SRR060687 9_0-2h_embryos_total |
V047 embryo |
M027 male body |
SRR060657 140_testes_total |
SRR060669 160x9_females_carcasses_total |
SRR060683 160_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AATCAACCACACGATGCAGAA............................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................ACGTCAAACAAGCAGCGCAACA..... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................GTACAGCTGGATGCAAATGTGCTT...................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....AACCACACGATGCAGACAGTACAGCT...................................................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................TACAGCTGGATGCAAATGTGCCT...................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ACAAACAGCTTTTGGTTTCCC................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GAGAAGACAACAGCGACGTCAA.................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AATCAACCACACGATGCAGAAAGTACA......................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...CAACCACACGATGCAGAAAGT............................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................GTACAGCTGGATGCAAATGTGCTTGAA................................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCGACGTCGGACAGGCAGCG.......... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AAGTACAGCTGGATGCAAATGT.......................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GAAGAAACTCCAGAGAAGACAAC............................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................GTACAGCTGGATGCAAATGT.......................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................CCATGGTATATCTCCTTCT................................................................. | 19 | 3 | 18 | 0.17 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| .........CAAGTTGCAGAAAGTGCAGCT...................................................................................................................................... | 21 | 3 | 18 | 0.11 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................ATATCTCCTTCTCAAAGTGA......................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TTAGTTGGTGTGCTACGTCTTTCATGTCGACCTACGTTTACACGGACTTCCATACAATGTACTGTTTGTCGAAAATTAAAGGGACGATTTAGAGGAAGAGTTAGACTTAATGTCTTTGAGGTCTCTTCTGTTGTCGCTGCAGTTTGTTCGTCGCGTTGTTAGCG
**************************************************....................((...(((((.........................)))))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060688 160_ovaries_total |
M047 female body |
SRR060676 9xArg_ovaries_total |
SRR060663 160_0-2h_embryos_total |
M027 male body |
SRR060672 9x160_females_carcasses_total |
SRR060683 160_testes_total |
SRR060666 160_males_carcasses_total |
SRR060678 9x140_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................CTGCAGTTTGTTCGTCGCGTT....... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TAAAGGGACGATTTAGAGGAA................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............ACGTCTTTCATGTCGACCTAC................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AGTCTCCTCTGTTGTCGC........................... | 18 | 2 | 3 | 0.67 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................CTGATCGTCGCGTTATTAGC. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CAGTCTCCTCTGTTGTCGC........................... | 19 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| ..........................................................................................................................CTCCTCTGTTGTCGCAGAAG...................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................GAGTTAGACATAATGTGT................................................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................GGGACGCTGTAGAGGGAGA................................................................. | 19 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:7303212-7303375 + | dvi_13426 | AATCAACCACACGATGCAGAAAGTACAGCTGGATGCAAATGTGCCTGAAGGTATGTTACATGACAAACAGCTTTTAATTTC-CCTGCTAAATCTCC---TTCTCAATCTGAATTACAGAAAC------TCCAGAGAAGAC---------------------------------------A---ACAGCGACG------TCAAACAAGCAGCGCAACAATCGC |
| droMoj3 | scaffold_6482:2634855-2635046 - | AGTTACCGAAACACTTCAGCAAGTACAGCTGGAAGCCCGTG---CAAAAGGTATATAACTTGACCAATA---TCTCACTACTT---TAACTTAACCTGTTTTCC-AAT-----CACAGCAAC------ATTGGAGGAGAATACAGAGCTGTCAGCAGAAGCAACGTCAACTGTAACAACAACCA---CATCA------GCAAATAAACAACGCAACAATCGC | |
| droGri2 | scaffold_14853:8892072-8892198 + | ACAATTC---------------------------------------------------------T---CAATTCACACAGCTA---CTAATTATCT---TATTCAAAT-----TGCAGCAGCAGCAGCAGCCGAGACGAC---------------------AACAACAACAATAACAATAGCAG---CGACGACAAATAAGCAGCAGCAGCGCAACAATCGC | |
| droAna3 | scaffold_13117:1890747-1890787 + | AGGATGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------G---A---CGACG---CCGCGGCAGCAGCAGGCCAAGGGTCGC | |
| droKik1 | scf7180000302577:1662424-1662460 - | GAC--------------------------------------------------------------------------------------------------------------------------GAC---------------------------------------------------G---A---CGACG------ACGAGGCAACAGCAAAAGGGTCGC | |
| droFic1 | scf7180000454116:203765-203796 - | GGC--------------------------------------------------------------------------------------------------------------------------AGC---------------------------------------------------AG-----------------CCTCATCAGCAGCAGAAGGGTCGC | |
| droEle1 | scf7180000491087:854610-854611 + | AA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000778730:17719-17767 + | AGAGAAAGACGC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------G---A---CGACGACGCGGCAACAGCAGCAACAAAAGGGTCGC | |
| droTak1 | scf7180000415324:191489-191580 - | GCAGCTA--AATGACACGGATTTAGCAGGTAAGCGCAAACGTCC---AAGTTATTCCACAAAA--AAGTAGTTTTGACAACT----CTAAATCTCC---TAATCAA-------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | X:21378636-21378667 - | GAC--------------------------------------------------------------------------------------------------------------------------AGC---------------------------------------------------AG-----------------TCACAACAACAACAGAAGGGTCGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/17/2015 at 04:24 AM