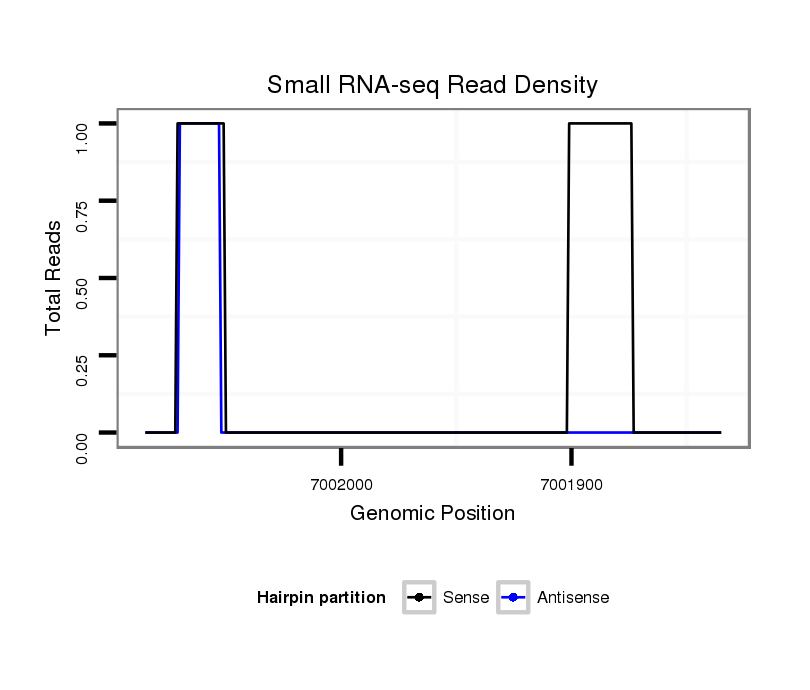
ID:dvi_13381 |
Coordinate:scaffold_12970:7001885-7002035 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
No conservation details. |
intron [Dvir\GJ18824-in]; exon [dvir_GLEANR_3644:2]; CDS [Dvir\GJ18824-cds]; intron [Dvir\GJ18824-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------########################################---------- GGTCCAGTTACGGAAACTCTTAACGAGACGTTTCTATGCTTGCCATACTATGTTCTCTTATCGCCCGATCGCTTTTATGCCAGATAAGATTTCGCTCATATATTTTGAGAAGCATGTAAATTCTCGTTGAAATAACTTCCTCGAAGGCCAAAGCACCGTTTTTCCTATAGAGCTTATATTATGTTATGTTCATATGATAAGAACATAATGGAGCTCATACTATATGCCAAGTTTAATGAGAGTATCTTAAA **************************************************.....(((((((((..((......))..))..)))))))....((((.((((....(((((((.(((....(((((..(((.....)))..)).))).....)))..))))))).))))))))((((..((((.......))))..)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060676 9xArg_ovaries_total |
SRR060660 Argentina_ovaries_total |
V116 male body |
SRR060664 9_males_carcasses_total |
M027 male body |
SRR060662 9x160_0-2h_embryos_total |
SRR060671 9x160_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................TATGTTCATATGATAAGAACATAATGGA....................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............AACTCTTAACGAGACGTTTCT........................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................ACTATATGGAAAGTCTAATGA............ | 21 | 3 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................GAGAGGTTTCTCTGCGTGC................................................................................................................................................................................................................ | 19 | 3 | 16 | 0.25 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| ...................................................................................................................................................CCAAAGCAGCGTATTTCTTAT................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GAACGCCAAGGCACCGGTT.......................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TTGCCTATAGAACTTATA......................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CCAGGTCAATGCCTTTGAGAATTGCTCTGCAAAGATACGAACGGTATGATACAAGAGAATAGCGGGCTAGCGAAAATACGGTCTATTCTAAAGCGAGTATATAAAACTCTTCGTACATTTAAGAGCAACTTTATTGAAGGAGCTTCCGGTTTCGTGGCAAAAAGGATATCTCGAATATAATACAATACAAGTATACTATTCTTGTATTACCTCGAGTATGATATACGGTTCAAATTACTCTCATAGAATTT
**************************************************.....(((((((((..((......))..))..)))))))....((((.((((....(((((((.(((....(((((..(((.....)))..)).))).....)))..))))))).))))))))((((..((((.......))))..)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060683 160_testes_total |
SRR060664 9_males_carcasses_total |
SRR060671 9x160_males_carcasses_total |
GSM1528803 follicle cells |
M047 female body |
SRR060681 Argx9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............TGAGAATTGCTCTGCAAA.......................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................AGCTTCCGGTTTCATCACAA........................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................TTCTAAAGCTAGTATATAG................................................................................................................................................... | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................ATACAAGTCGACTATTCTTG............................................... | 20 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................TGAGCTACTGTTCAAATTAC............. | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................GTTGGTGCCAAAAAGGAT.................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................CAACTTCCCGTTTCGTGGC............................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:7001835-7002085 - | dvi_13381 | GGTCCAGTTACGGAAACTCTTAACGAGACGTTTCTATGCTTGCCATACTATGTTCTCTTATCGCCCGATCGCTTTTATGCCAGATAAGATTTCGCTCATATATTTTGAGAAGCATGTAAATTCTCGTTGAAATAACTTCCTCGAAGGCCAAAGCACCGTTTTTCCTATAGAGCTTATATTATGTTATGTTCATATGATAAGAACATAATGGAGCTCATACTATATGCCAAGTTTAATGAGAGTATCTTAAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/17/2015 at 03:54 AM