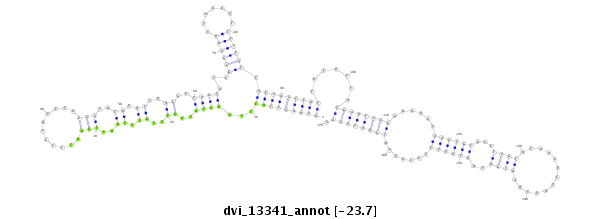
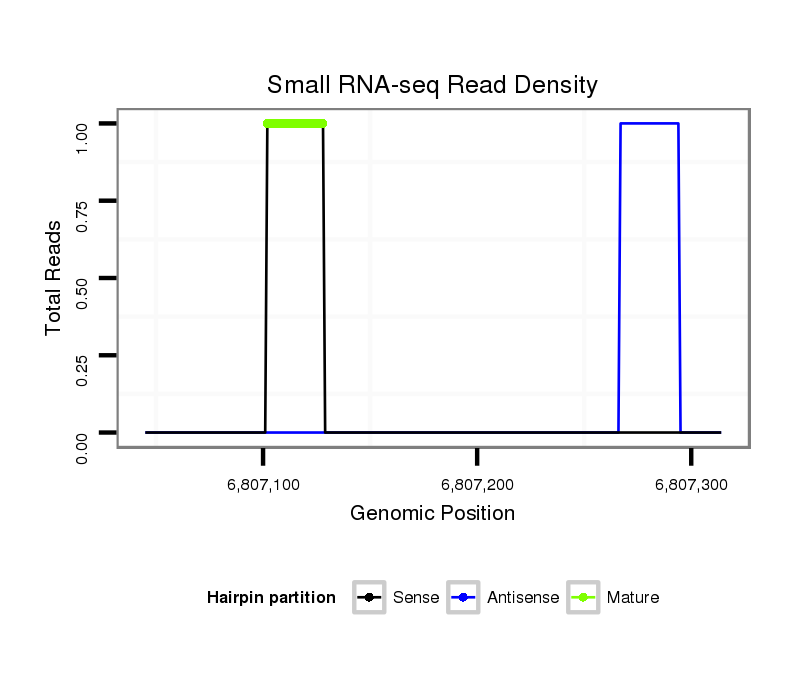
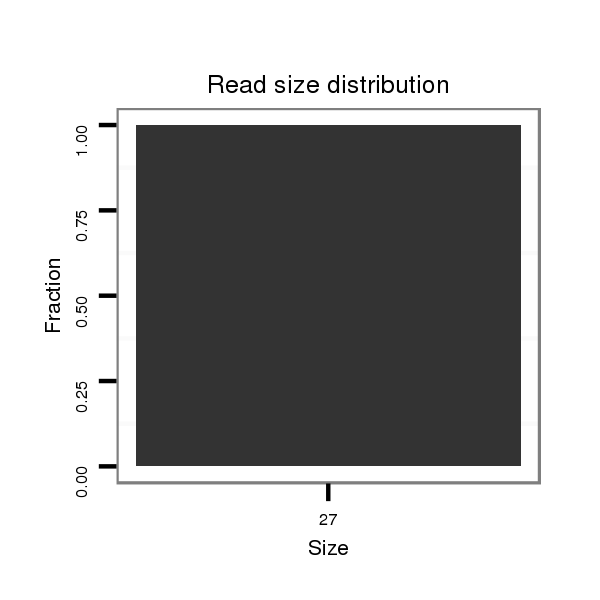
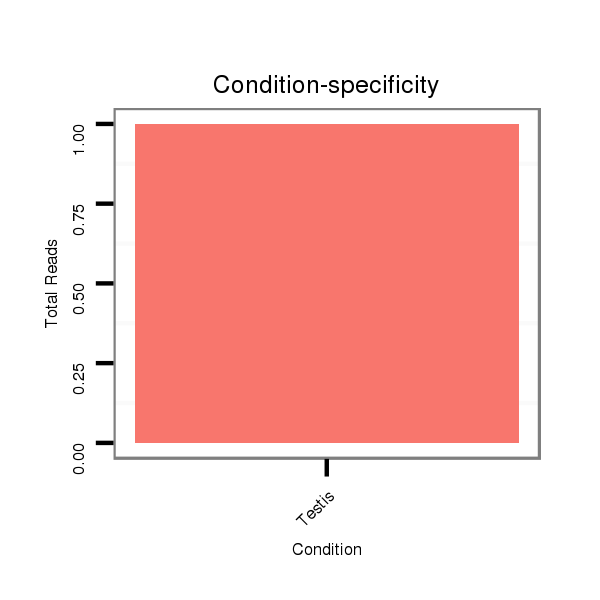
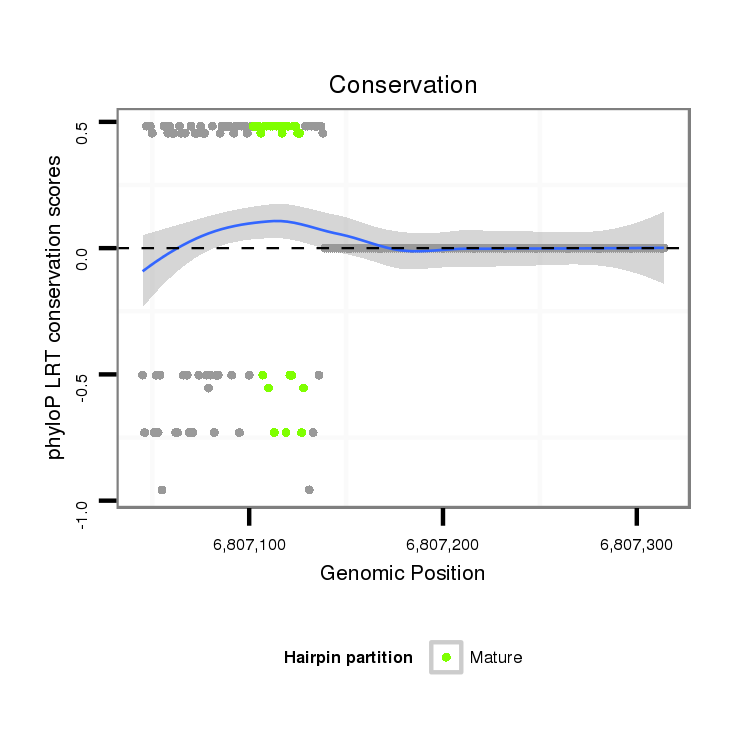
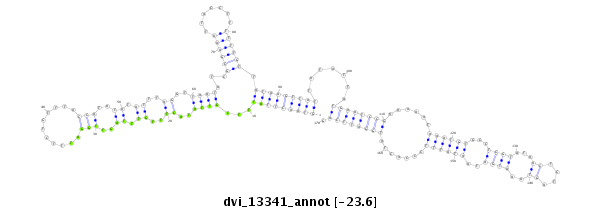
ID:dvi_13341 |
Coordinate:scaffold_12970:6807095-6807264 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -23.7 | -23.6 | -23.0 |
 |
 |
 |
exon [dvir_GLEANR_4217:3]; exon [dvir_GLEANR_4217:4]; CDS [Dvir\GJ19326-cds]; CDS [Dvir\GJ19326-cds]; intron [Dvir\GJ19326-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATATTCAAACGTTCTGGAAACGCGATACGAACCAATTCATGAAAATTGTTGTAACTTATTTCGTTATACTTTGTTTAGTTGGATTTCTGTTTACCATATATATTTAGGTTAATATCGCGAAATTACCTTTTTTGTTATAAGTTATTATATTATGAATCCGAAAAATAAATGTGCCTCGTATAACTCTAGTAAATGATAATATTTCCCACTATGGATTCAGCTATATACGCTGAAGTATCTATGGATAACAACAACAACGACAAGCCAAGA **************************************************(((((((((...((((..((..((.((..(((...........)))..)).))..))..))))...((((((.......)))))).))))))))).......((((((((......((((((...((((.............))))..)))))).......)))))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
V116 male body |
SRR060665 9_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................ATTTCGTTATACTTTGTTTAGTTGGAT.......................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................GCTGAAGTGTCTATGGGTA....................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................AATGGTAACATTTCCCACTGT.......................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................GTGGTGGAACTTATTTCGTT............................................................................................................................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................CTATTCGTGAAAATTGTT............................................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................GTTTAGGTGGATTACTGTG................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................GAGGTTAATATCACGAGA.................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TATAAGTTTGCAAGACCTTTGCGCTATGCTTGGTTAAGTACTTTTAACAACATTGAATAAAGCAATATGAAACAAATCAACCTAAAGACAAATGGTATATATAAATCCAATTATAGCGCTTTAATGGAAAAAACAATATTCAATAATATAATACTTAGGCTTTTTATTTACACGGAGCATATTGAGATCATTTACTATTATAAAGGGTGATACCTAAGTCGATATATGCGACTTCATAGATACCTATTGTTGTTGTTGCTGTTCGGTTCT
**************************************************(((((((((...((((..((..((.((..(((...........)))..)).))..))..))))...((((((.......)))))).))))))))).......((((((((......((((((...((((.............))))..)))))).......)))))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060681 Argx9_testes_total |
V053 head |
SRR060689 160x9_testes_total |
SRR060665 9_females_carcasses_total |
M061 embryo |
SRR060668 160x9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................TATATGCGACTTCATAGATACCTATTGT.................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............AGACCTTTGCGCTATGCTCGGTTAA......................................................................................................................................................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................AAGCAGAATGAAACAAGTCA............................................................................................................................................................................................... | 20 | 3 | 17 | 0.12 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................TTACCATTGTAAAGGGTGACA.......................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................GCTTTTTATTGCGACGGAGC............................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................TTGAGTAGTTTTAACAACA.......................................................................................................................................................................................................................... | 19 | 2 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TGGTACAAGCAATATTCAA............................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................TACATACGCTTTTTATTTACAA................................................................................................. | 22 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TAACAATATTCAGTAATATT........................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:6807045-6807314 + | dvi_13341 | ATATTCAAACGTTCTGGAAACGCGATACGAACCAATTCATGAAAATTGTTGTAACTTATTTCGTTATACTTTGTTTAGTTGGATTTCTGTTTACCATATATATTTAGGTTAATATCGCGAAATTACCTTTTTTGTTATAAGTTATTATATTATGAATCCGAAAAATAAATGTGCCTCGTATAACTCTAGTAAATGATAATATTTCCCACTATGGATTCAGCTATATACGCTGAAGTATCTATGGATAACAACAACAACGACAAGCCAAGA |
| droMoj3 | scaffold_6473:12248582-12248675 + | GGATTCCGCTCTTCTGGCCACACACTCCGGACCGTCTAGCGAAAATCGTTTTAACCTATTTCATTTTAATTTGTGTGATTGGCATTGTTTTCAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/17/2015 at 03:34 AM