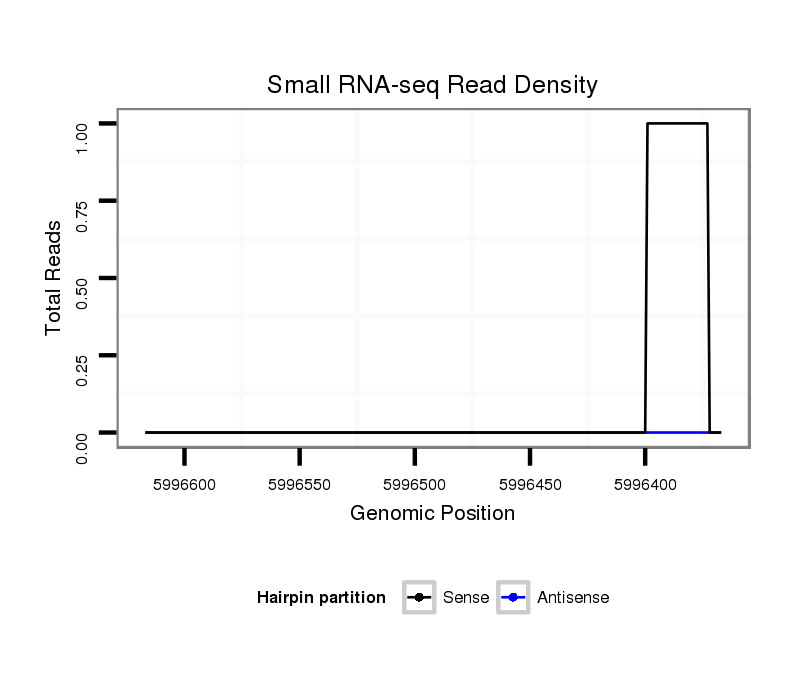
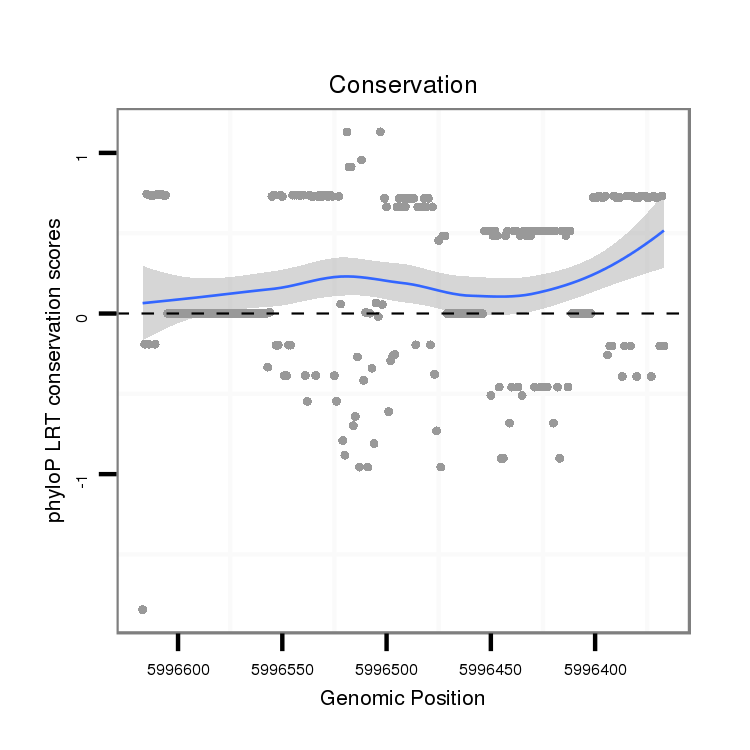
ID:dvi_13276 |
Coordinate:scaffold_12970:5996417-5996567 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_3680:2]; CDS [Dvir\GJ18861-cds]; intron [Dvir\GJ18861-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GTTTGGGATTCGAACTCGTGAGAAAACGCAAGATTGCATATATTACGGCACGTTTGAACCGGCAACTGCGCCTTAATTGGTGCACACAAGTCAGCCCAACCCTTGAGTGGTATAACTGGGAACAGAGTATTGCGCAGTCGTGGGATATACGCATTTATTTGAATTTAAGTCTCGCTTGAATGATCTCATTTCTCCTTGTAGATGTTTCGATCCGTTGGCAGGTAAGACCCGCAACAGGGATTGGCTGCAAA **************************************************........((.((.((((((((((((((((.((..........))))))...))))).(((((..(((....))).)))))))))))).)).))...((((........(((((((((((...))))))))....)))........)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060671 9x160_males_carcasses_total |
SRR060673 9_ovaries_total |
SRR060674 9x140_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060659 Argentina_testes_total |
M027 male body |
SRR060681 Argx9_testes_total |
M028 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................AACTGGGAACAGAGTATGGCGCAGTC................................................................................................................ | 26 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GATGTTTCGATCCGTTGGCAGGTGA.......................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................AACTGGGAACAGATTATGGCGCAGTC................................................................................................................ | 26 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TATAACTGGGAACAGAGTATTACGC.................................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................TAAGTCTCGTTTGAATGATCTCATT............................................................. | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......ATTCGAGCTCGTGAGAAAACGCA............................................................................................................................................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................CAGGTAAGACCCGCAACAGGGATTGGC...... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................TGGGAACAGAGTATGGCGCAGTC................................................................................................................ | 23 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................TGAATGATCGGATATCTCCT....................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................CCGTGGGCTGGTAAGAGCC..................... | 19 | 3 | 15 | 0.20 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ..................................................................................................................................TGCCCAGTCGTGGGGGATA...................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................CGAAACAGGGATTGACGGC... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CAAACCCTAAGCTTGAGCACTCTTTTGCGTTCTAACGTATATAATGCCGTGCAAACTTGGCCGTTGACGCGGAATTAACCACGTGTGTTCAGTCGGGTTGGGAACTCACCATATTGACCCTTGTCTCATAACGCGTCAGCACCCTATATGCGTAAATAAACTTAAATTCAGAGCGAACTTACTAGAGTAAAGAGGAACATCTACAAAGCTAGGCAACCGTCCATTCTGGGCGTTGTCCCTAACCGACGTTT
**************************************************........((.((.((((((((((((((((.((..........))))))...))))).(((((..(((....))).)))))))))))).)).))...((((........(((((((((((...))))))))....)))........)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR1106729 mixed whole adult body |
SRR1106727 larvae |
SRR1106728 larvae |
V116 male body |
SRR060659 Argentina_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
M047 female body |
SRR060667 160_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
M027 male body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060687 9_0-2h_embryos_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................TCGGGTTGGGAACCC................................................................................................................................................ | 15 | 1 | 10 | 46.80 | 468 | 336 | 67 | 65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................CTTGGGCATTGAGGCGGAA................................................................................................................................................................................. | 19 | 3 | 5 | 1.60 | 8 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CAAAGCTAGGCTCCTGTCC............................. | 19 | 3 | 8 | 0.50 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................GTGTTCAGTAGGTTTGGGA.................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CTCTAAGCTTGAGTATTCTT................................................................................................................................................................................................................................... | 20 | 3 | 9 | 0.33 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...ACTCTAAGCTTGAGTATTCTT................................................................................................................................................................................................................................... | 21 | 3 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ....................................................................................GTGTTCACTCGCGTTGGGAG................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................GCACATAATGCCGTGCAA..................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......CTAAGCTTGAGTATTCTT................................................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CAAAGCTAGGCTCCTGTC.............................. | 18 | 3 | 20 | 0.20 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................GGTGTTCAGTAGGTTTGGGA.................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TGTCGGGTTGGGAACCC................................................................................................................................................ | 17 | 2 | 16 | 0.19 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TGTCGGGTTGGGAAC.................................................................................................................................................. | 15 | 1 | 18 | 0.17 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TTGTCGGGTTGGGAACTC................................................................................................................................................ | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TCGGGTTGGGAACGC................................................................................................................................................ | 15 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCAGAGACAACTAACTAGAG................................................................ | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................TCGGGTTGGGAACTAGC.............................................................................................................................................. | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TTGTCGGGTTGGGAACT................................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:5996367-5996617 - | dvi_13276 | GTTTGGGATTCGAACTCGTGAGAAAACGCAAGATTGCATATATTACGGCACGTTTGAACCGGCAACTGCGCCTTAATTGGTGCACACAAGTCAGCCCAACCCTTGAGTGGTATAACTGGGAACAGAGTATTGCGCAGTCGTGGGATATACGCATTTATTTGAATTTAAGTCTCGCTTGAATGATCTCATTTCTCCTTGTAGATGTTTCGATCCGTTGGCAGGTAAGACCCGCAACAGGGATTGGCTGCAAA |
| droMoj3 | scaffold_6359:2045428-2045481 - | A-----------------------------------------------------------AC---------------------------------CAGACCTGCTAATTGCGCAAATGCGTGCAGAGTATTGCGCAGTCGTTGCAT--------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15110:20912445-20912522 - | T-----------------------------------------------------------GG---------------------------------TATA-------------TCACTGGTAACAGAGTATTACGCAGTTGG-----------------------TTATGTCCGCCTGAAACGTTCTCACTTTTTTTTTTGCATGCT--------------------------------------------- | |
| droBip1 | scf7180000396696:1800-1856 + | C-----------------------------------------------------------GGCAGTTGATTTTTAATTTCTGCCCACAAGTCCCCCCAACCATCCAACCATCTAAC--------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000780072:25100-25136 + | T-----------------------------------------------------------T-----------------------------------------------------------------------------------------------------------------------------------------------------------GGCAGGTTGAACCCTTAATAGTGATTGGATGCGAG | |
| droBia1 | scf7180000302069:380955-380966 + | GCTCGGAATTCG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
Generated: 05/16/2015 at 10:27 PM