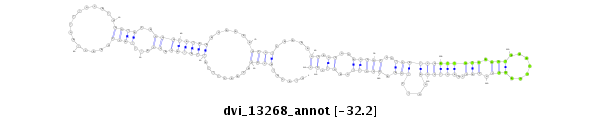
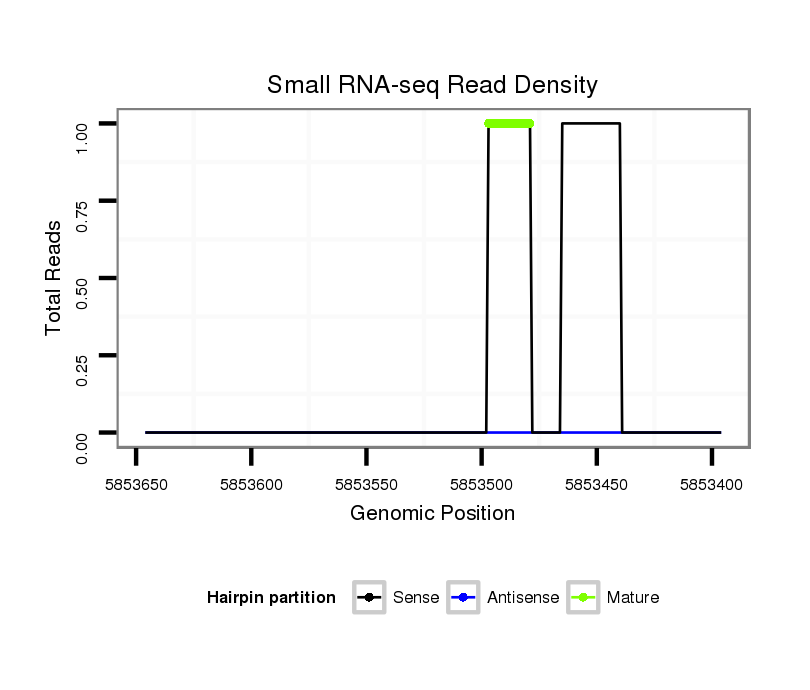
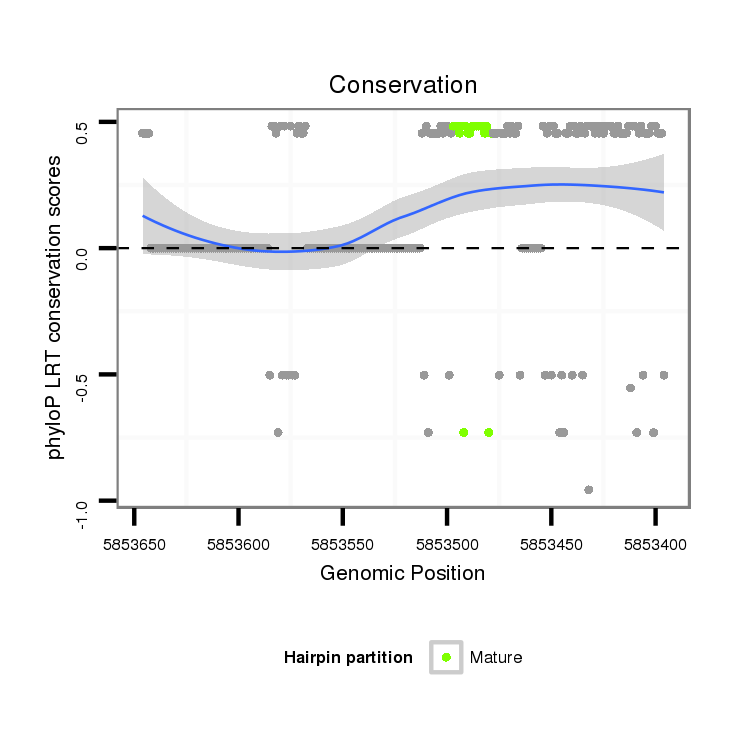
ID:dvi_13268 |
Coordinate:scaffold_12970:5853446-5853596 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.9 | -31.8 | -31.7 |
 |
 |
 |
exon [dvir_GLEANR_3682:1]; CDS [Dvir\GJ18863-cds]; intron [Dvir\GJ18863-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGCGTGGTTACTCAGCGGGAAGTCGCAGAGAGTGCCTCTCAGCGTCTAGCGTACGTACGCACAACTTTATTATACACCATTTACCGAACTAGTTTGCATGGGTACAGGGTATAAACAAAAGCGTCAAAAGCAGCGGAGCGCCGTCATCGATTGTGAGGTAAATTGAGCCGCACCGAGTCGAGGTCTGAAGCGTTGAGTCGCGTCGAGTCGAGCTGAAGTGTCATTTGCACGCCTATTTCTTGGCTGTGCCC **************************************************......((((.........((((((.((...((((...............))))..))))))))......)))).....((.((..(((((..((((((((((((.(((.......))).)))..))))))....))).)))))..)).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060657 140_testes_total |
SRR060658 140_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
V053 head |
SRR060668 160x9_males_carcasses_total |
M047 female body |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................GGTCTGAAGCGTTGAGTCGCGTCGAG............................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............CGGGAACTCGCAGACGGTGCCT...................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................AGCCGCACCTAGTTGAGGT................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ATTGTGAGGTAAATTGAGC................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTACGTACCGACTACTTTAT..................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................TGTCAAAGGCAGTGGAGCGCC............................................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................CGCTCAGCGTCTAGC......................................................................................................................................................................................................... | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CCGCACCAATGAGTCGCCCTTCAGCGTCTCTCACGGAGAGTCGCAGATCGCATGCATGCGTGTTGAAATAATATGTGGTAAATGGCTTGATCAAACGTACCCATGTCCCATATTTGTTTTCGCAGTTTTCGTCGCCTCGCGGCAGTAGCTAACACTCCATTTAACTCGGCGTGGCTCAGCTCCAGACTTCGCAACTCAGCGCAGCTCAGCTCGACTTCACAGTAAACGTGCGGATAAAGAACCGACACGGG
**************************************************......((((.........((((((.((...((((...............))))..))))))))......)))).....((.((..(((((..((((((((((((.(((.......))).)))..))))))....))).)))))..)).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
M027 male body |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|
| ............................................................................................................CATATTCCTTTTCGCAGTT............................................................................................................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 |
| .........................................................................................................................GCAGTGTTCGTGGCCTCG................................................................................................................ | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ....................TCAGCGTCTATCACGGAAAT................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................GGCTTCACAGTTAGCGTGC.................... | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 |
| ....................................AGAATCACAGTTCGCATGC.................................................................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 |
| ....................GCAGCGTCTATCACGGAGAT................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:5853396-5853646 - | dvi_13268 | GG--------CGTGGTTACTCAGCGGGAAGTCGCAGAGAGTGCCTCTCAGCGTCTAGCGTACGTACGCACAACTTTATTATACACCATTTACCGAACTAGTTTGCATGGGTACAGGGTATAAACAAAAGCGTCAAAAGCAGCGGAGCGCCGTCATCGATTGTGAGGTAAATTGAGCCGCACCGAGTCGAGGTCTGAAGCGTTGAGTCGCGTCGAGTCGAGCTGAAGTGTCATTTGCACGCCTATTTCTTGGCTGTGCCC |
| droMoj3 | scaffold_6473:15782212-15782348 + | GGGTATGGGGCG---------------------------------------------------------TAACGTCACCACGCACCA-------------------------------------------------------GAATCGCCGTCATTGATTGTTAGGTAAATTGATCCGCGCCGAGTCGAA----------TCGAATCGAAGCGAATCGAACTCAAGTGTCATTTGCACGCCTTTTGCTCGGCTTTGCCT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 10:15 PM