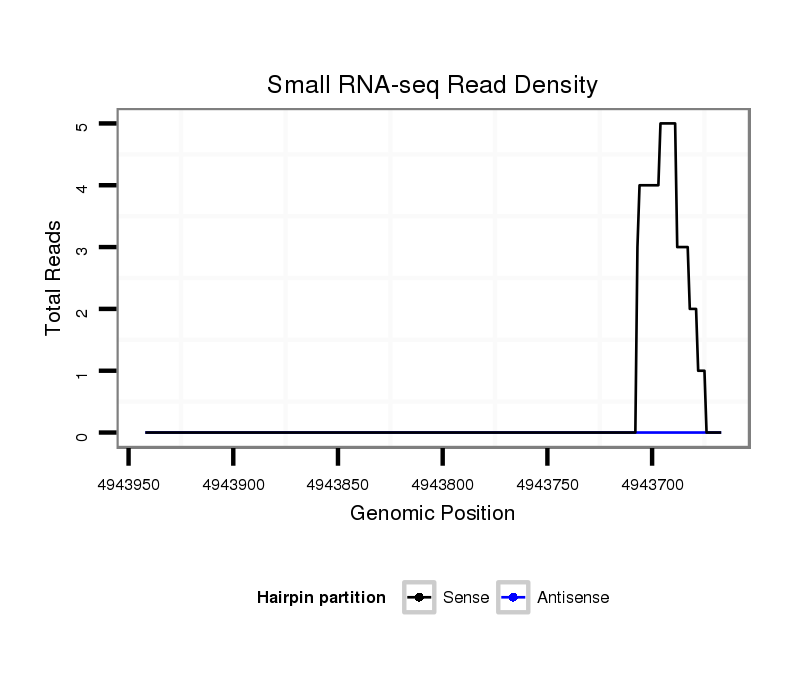
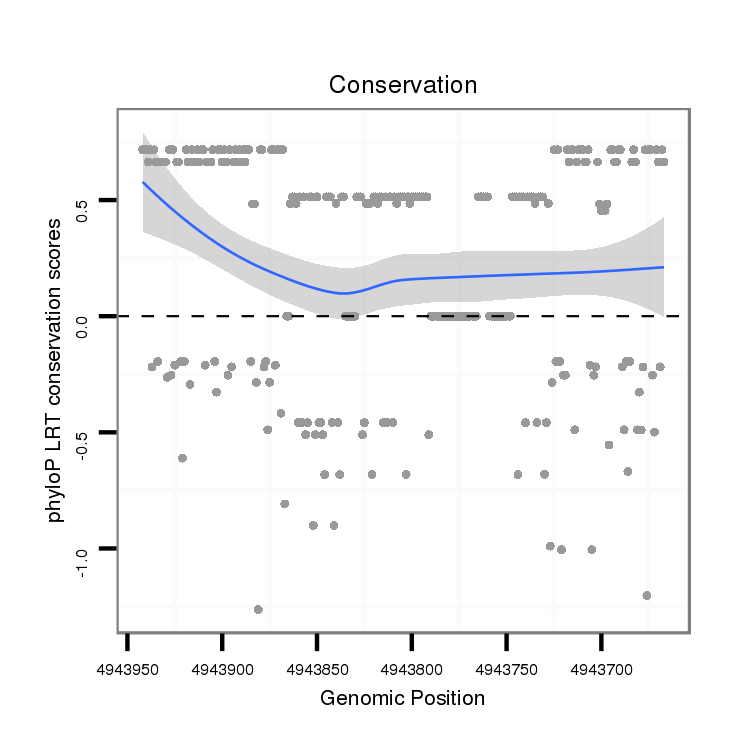
ID:dvi_13160 |
Coordinate:scaffold_12970:4943717-4943892 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ18914-cds]; CDS [Dvir\GJ18914-cds]; exon [dvir_GLEANR_3728:1]; exon [dvir_GLEANR_3728:2]; intron [Dvir\GJ18914-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATTGAGAGCCCCCATTAAGGGCCTGCAGCTGAAACGCTCCTTGACTTACAGTGAGTAGATTCAATGGTATATAAAGTACATCGAGAACATGTTGGACAATGGGCCTACCTATCTATTCGCGCTTCTTTTATATTCATATTTCATTTATATAACGTAACAAACGCACACACACGTATTATATAATAATAGTAAATGATAGATACATATGTTAATGCATTCTCTTTAGATCCAAAGCAATTGCAGCCATTTGCTTTGGGCGAGGCTGTCAAAAAGGAG **************************************************((((((((((.....(((.(((.....((((.......))))......))).))).....))))))))))((......(((((..((...((((((((....(((.....)))..........((((((...))))))))))))))...))..))))).....))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060683 160_testes_total |
SRR060689 160x9_testes_total |
SRR060657 140_testes_total |
SRR060665 9_females_carcasses_total |
SRR1106729 mixed whole adult body |
SRR060671 9x160_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
V116 male body |
SRR060658 140_ovaries_total |
GSM1528803 follicle cells |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................................AATTGCAGCCATTTGCTTT...................... | 19 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................GACGAGTCTGTCCAAAAGGA. | 20 | 3 | 4 | 1.50 | 6 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................................ATTGCAGCCATTTGCTTTGGGCGAGGCT............ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................AATTGCAGCCATTTGCTTTGGGCGA................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................TACCTATCTATTCGCGCTCCTTTTA.................................................................................................................................................. | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................TTTGCTTTGGGCGAGGCTGTCA........ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................ATATTGGACAATGGG............................................................................................................................................................................. | 15 | 1 | 15 | 0.33 | 5 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................ACAATAAGAGTAAATGAGAGAT........................................................................... | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................TTCAAATAACGTAACAAATGC................................................................................................................ | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................ATTCAAATAACGTAACAAATG................................................................................................................. | 21 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TCAAATAACGTAACAAATGCA............................................................................................................... | 21 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................ATTCATTTTAGATCCAA............................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TAACTCTCGGGGGTAATTCCCGGACGTCGACTTTGCGAGGAACTGAATGTCACTCATCTAAGTTACCATATATTTCATGTAGCTCTTGTACAACCTGTTACCCGGATGGATAGATAAGCGCGAAGAAAATATAAGTATAAAGTAAATATATTGCATTGTTTGCGTGTGTGTGCATAATATATTATTATCATTTACTATCTATGTATACAATTACGTAAGAGAAATCTAGGTTTCGTTAACGTCGGTAAACGAAACCCGCTCCGACAGTTTTTCCTC
**************************************************((((((((((.....(((.(((.....((((.......))))......))).))).....))))))))))((......(((((..((...((((((((....(((.....)))..........((((((...))))))))))))))...))..))))).....))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060666 160_males_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060667 160_females_carcasses_total |
M028 head |
M047 female body |
SRR060668 160x9_males_carcasses_total |
M027 male body |
V053 head |
SRR060684 140x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR1106714 embryo_2-4h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................GTTCTTTACGTGTGTGTGCATA.................................................................................................... | 22 | 3 | 7 | 1.43 | 10 | 6 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TTCTTTACGTGTGTGTGCATA.................................................................................................... | 21 | 2 | 2 | 1.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGTTCTTTACGTGTGTGTGCATA.................................................................................................... | 23 | 3 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCTTTACGTGTGTGTGCATA.................................................................................................... | 20 | 2 | 7 | 0.86 | 6 | 1 | 1 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................TTTACGTGTGTGTGCATA.................................................................................................... | 18 | 1 | 10 | 0.70 | 7 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................CCCCGATCAGACAGTTTTTC... | 20 | 3 | 9 | 0.33 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ACAGTTCCGTCAGAGAAATC.................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GGGATAGATTAGCGCGATG....................................................................................................................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TAATTACGTAAGAGAAATA.................................................. | 19 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TCGACTTTGGGAAGAAGTGA...................................................................................................................................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TCGGCTTTGGGAGGAAGTG....................................................................................................................................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AGCTCTCGGGGGGAATCC................................................................................................................................................................................................................................................................. | 18 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................GGTTTCATTAAAGTCTGTAAA........................... | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................CTTTACGTGTGTGTGCATA.................................................................................................... | 19 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................GAACTTGCGAGCAACTGAA..................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................AGGAACTGAATGCAGCTC............................................................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................GGAGCCCTTGCACAACCT.................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:4943667-4943942 - | dvi_13160 | ATTGAGAGCCCCCATTAAGGGCCTGCAGCTGAAACGCTCCTTGACTTACAGTGAGTAGATTCAAT----GGTAT-------------------------------------------------ATAAAGTACATCGAGAACATGTTGGACAATGGGCC-TACCTATCTATTCGCGCTT-------C--TTTTATATTC-ATATTTCATTTATATAACGTAACAAACGCACACACACGTATTATATAATAATAGTAAATGATAGATACATAT-GTTAATGC--------------ATT-CTCTTTAGATCCAAAGCAATTGCAGCCATTTGCTTTGGGCGAGG---------CTGTCAAAAAGGAG |
| droMoj3 | scaffold_6473:4427053-4427255 - | ATTGAAAGCCCCCTTCAAGGGGCTGCAGCTGAAACGCTCATTGACCTACAGTGAGTAGATCGAATTGATGGCATTGTTGTTCACTGAATTGATGCACATATATCCATAATCGATTAATCGAATATATAC-----------------------------------------------------------------------------------------------------------------------------------------------------------------ACCTTCT-CTCGCCAGATCCAAAGCAAGCACAGCCAATTGCTTTGGTCGAGGTCTCTGAGGAGATTAAGCAGAAG | |
| droGri2 | scaffold_15081:397725-397994 + | ATTGAGAGTCCCCATTAGGGACTTGAAGCTGAAGCGCTTCTTGACTTTCAGTGAGTAA--CAAAATGCCACCATAATTGCTT---------------------------------ATATGTATGTATAC--CATCAAAATTATCATAATAAATACGTATTA------TATATGCGATTGAGATTTCAATTCTGTACTCAATATGTCATTTATATAT-------------------------ATATAA------------ATATATATATATAGCTAAGACATTTATATTTATCTCCTGTTTATTAGATGCAAAGCAGATGC------TTGCTTACATTGAGC---------CGGTGAAAAAGGAG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 08:39 PM