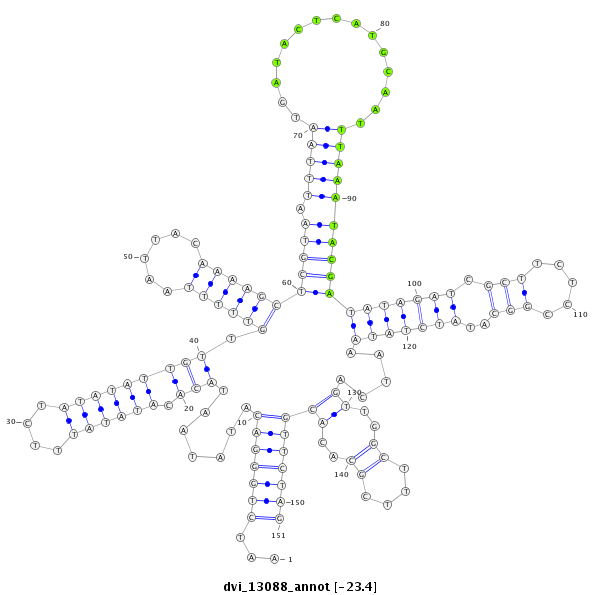
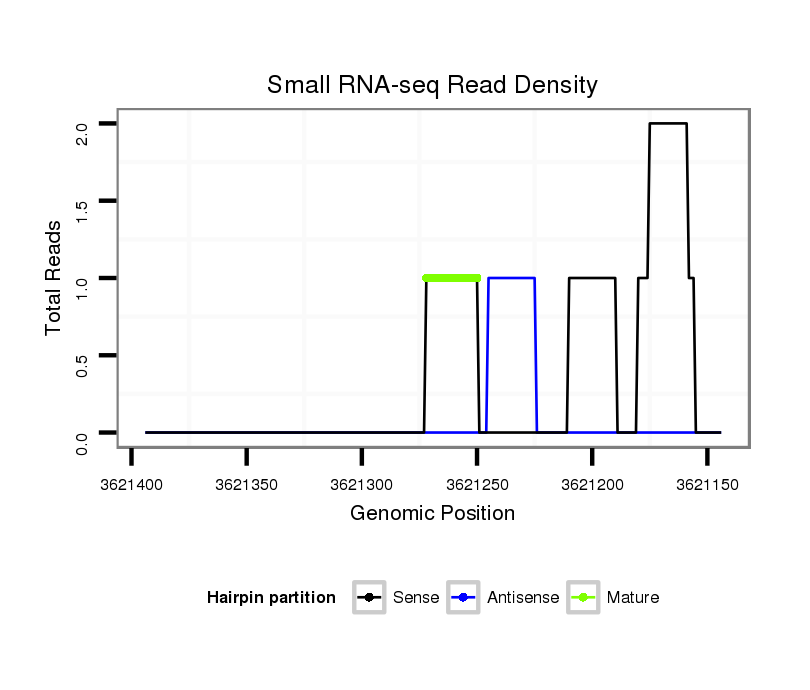
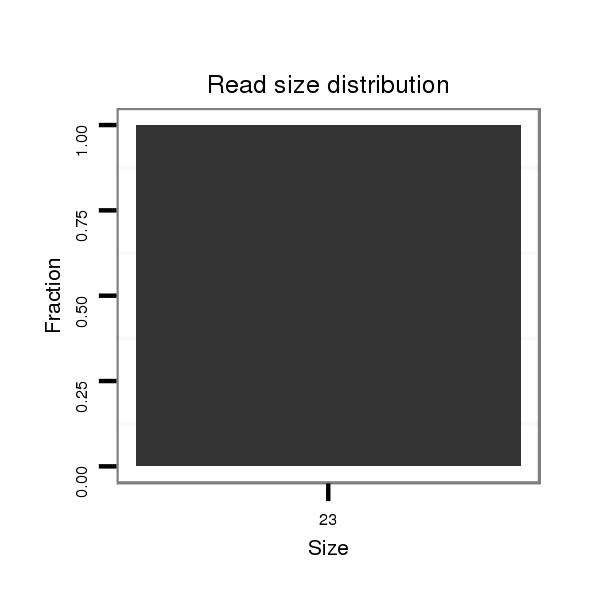
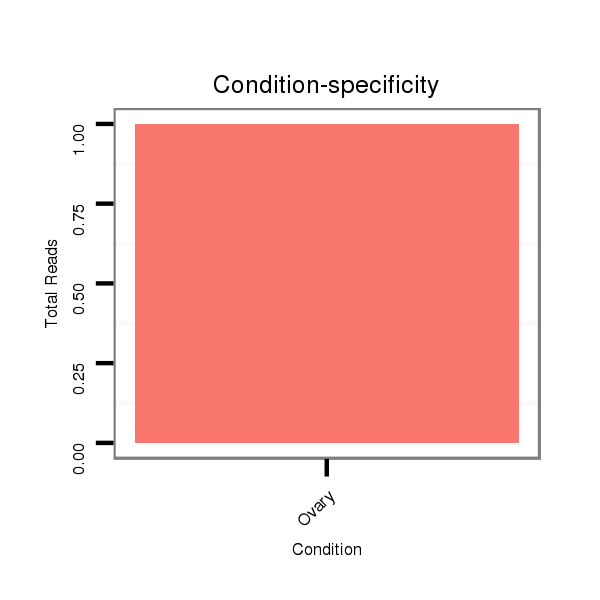
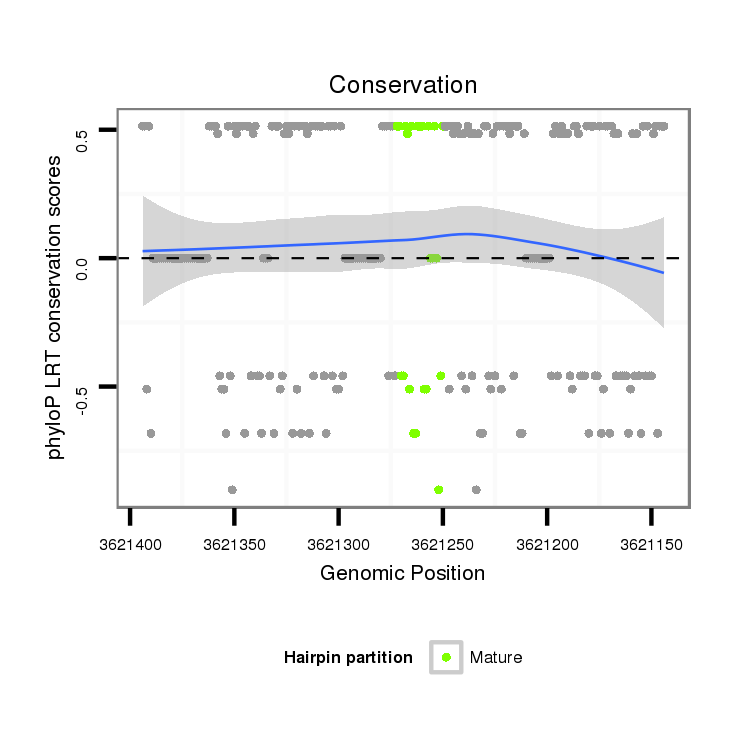
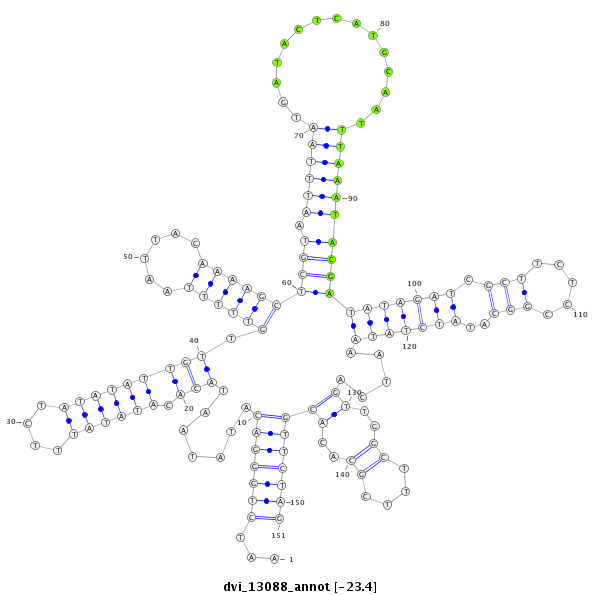
ID:dvi_13088 |
Coordinate:scaffold_12970:3621194-3621344 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -23.4 | -23.2 | -23.2 |
 |
 |
 |
exon [dvir_GLEANR_3772:4]; CDS [Dvir\GJ18958-cds]; intron [Dvir\GJ18958-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AATTGTAAGCGTCAAGCGAAGAACAGAAAAGTTAAACCTTGTCGTCTATTAATCTGGGACATATAATACACATATATTTCTATATATTGTTGTTTTTAATTACAAAAGCTCGTAATTTAATGATACTCATGCAATTTAAATACGATATAGATCGCTTCTCCGGCATATCTATAAATCAGTTGGCTTTCGCACACGTTCTAGACGCTAATGGAAATAAGGTTATTGTGCCGCGCCAGTCGGTCAATGAAAAT **************************************************...(((((((.......(((.((((((....))))))))).((((((......))))))(((((.(((((...............))))))))))(((((((.(((.....)))..))))))).....((..((....))..)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060681 Argx9_testes_total |
V116 male body |
SRR060660 Argentina_ovaries_total |
SRR060672 9x160_females_carcasses_total |
SRR060666 160_males_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060668 160x9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................TTACAAAAGCTCGTAGTTGAA................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................TTTCGCACACGTTCTAGACGC.............................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ATACTCATGCAATTTAAATACGA.......................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TTATTGTGCCGCGCCAGTCG............ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TAAGGTTATTGTGCCGCGCCAG............... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ATGCTAATGGTAATAAGGT............................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ATGCTAATGGTAATAAGGTTGT............................ | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................CAAAAGCTCGTAGTTGAA................................................................................................................................... | 18 | 2 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................AGAAGTTAAACCTTGGGGT.............................................................................................................................................................................................................. | 19 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................TTAAAAAGCTCGTAGTTTAA................................................................................................................................... | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........GTCAAACGAAGAACCGGAAA............................................................................................................................................................................................................................. | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................ACGTTCTAAACGCTACTGA........................................ | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................TAAAAGCTCGTAATTGAA................................................................................................................................... | 18 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................AAAAAGCTCGTAGTTTAA................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
TTAACATTCGCAGTTCGCTTCTTGTCTTTTCAATTTGGAACAGCAGATAATTAGACCCTGTATATTATGTGTATATAAAGATATATAACAACAAAAATTAATGTTTTCGAGCATTAAATTACTATGAGTACGTTAAATTTATGCTATATCTAGCGAAGAGGCCGTATAGATATTTAGTCAACCGAAAGCGTGTGCAAGATCTGCGATTACCTTTATTCCAATAACACGGCGCGGTCAGCCAGTTACTTTTA
**************************************************...(((((((.......(((.((((((....))))))))).((((((......))))))(((((.(((((...............))))))))))(((((((.(((.....)))..))))))).....((..((....))..)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
GSM1528803 follicle cells |
SRR060684 140x9_0-2h_embryos_total |
M061 embryo |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................CTAGCGAAGAGGCCGTATAGA................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................ATGAGTAAGTAAAATTTA.............................................................................................................. | 18 | 2 | 20 | 0.90 | 18 | 0 | 18 | 0 | 0 | 0 |
| ......................................................................................................................................................TAGCGACGTGGCCGTATA................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................CAACAAAAGTTAATGTTATCGA............................................................................................................................................. | 22 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................GCACGGCGCGCACAGCCAGT........ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:3621144-3621394 - | dvi_13088 | AATTGTAAGCGTCAAGCGAAGAACAGAAAAGTTAAACCTTGTCGTCTATTAATCTGGGACATATAATACACATATATTTCTATATATTG--------TTGTTTTTAATTACAAAAGCTCGTAATTTAATGATACTCATGCAATTTAAATACGATATAGATCGCTTCTCCGGCATATCTATAAATCAGTTGGCTTTCGCACACGTTCTAGACGCTAATGGAAATAAGGTTATTGTGCCGCGCCAGTCGGTCAATGAAAAT |
| droGri2 | scaffold_15203:10267228-10267423 + | AAATT---------------------------TAAACTAATTTCTCTATGAACCTAAT---CAGAAAGCACAGAAAGTTCGACATATCTAAATATGATTATAATC------------------TTTGATAATGTTCTTTAAATAAA--T-GAATAAAGATCACATCCCGGTAATGACCATTAATCAATTTTC------------CCTGGACGCCTATGAGGAGAAAATGTTTTTGTCATATATGCCATTTAGCGACAAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 05:54 PM