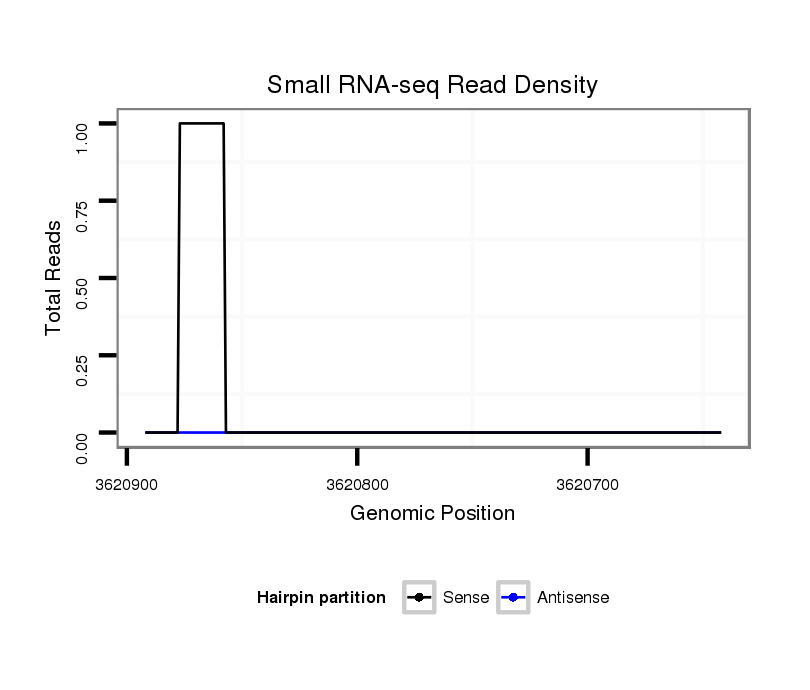

ID:dvi_13087 |
Coordinate:scaffold_12970:3620692-3620842 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ18958-cds]; exon [dvir_GLEANR_3772:5]; intron [Dvir\GJ18958-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTCAACCCGAATGGATATCTGTGGCGAGGATTACGCTGCCTCTTCGCCTGGTAAGTCTGCCATATACTAAAAAGGGCTAGTTGGTCACGACCAAGTGATTAGGATATATATTAATGTACATTAAGATTGTCATTTAGCTAGTTAGAAGAATAGAAGTAATATATTTTAATGCGAGGTGAAGGTAAAAGTAAAAGTTCGAGCTTTAATTAACTTACATAAACAGCTTTTCATATGTTTTTCTCTTTTTTTTT **************************************************...(((((......(((.............((((((((......)))))))).............))).....))))).((((((.((..(((((((..(((.......))).))))))))).))))))......................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060657 140_testes_total |
V047 embryo |
V053 head |
V116 male body |
SRR060666 160_males_carcasses_total |
SRR060655 9x160_testes_total |
M027 male body |
M028 head |
SRR060686 Argx9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060659 Argentina_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................TGTGGCGAGGATTGCGGTTC.................................................................................................................................................................................................................... | 20 | 3 | 9 | 1.22 | 11 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............TATCTGTGGCGAGGATTACG........................................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................TGTGGCGAGGAGTGCGCTTC.................................................................................................................................................................................................................... | 20 | 3 | 10 | 0.90 | 9 | 6 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................GTGGCGAGGAGTACGGTTC.................................................................................................................................................................................................................... | 19 | 3 | 9 | 0.33 | 3 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................TGTGGCGAGGAGTACGGTTC.................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................ACGATGCCTCCTCGCCGGGT....................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................CGTGTGGCGAGGATTGCG........................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GTGGCGAGGAGTGCGCTTC.................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.30 | 6 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................GTAACGCTGCCATATACTAC..................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................CGGTGGCGAGGATTACGA....................................................................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................AAGAGCTAGTCGGTCATGAC................................................................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................TGTGGCGAGGAGTGCGGTGC.................................................................................................................................................................................................................... | 20 | 3 | 13 | 0.15 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................CAAGTGGTTAAGATATATAT............................................................................................................................................ | 20 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................GTTATGATGCGAGGTGAAG...................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................GTGGCGAGGATTGCGGTTCC................................................................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CAGTTGGGCTTACCTATAGACACCGCTCCTAATGCGACGGAGAAGCGGACCATTCAGACGGTATATGATTTTTCCCGATCAACCAGTGCTGGTTCACTAATCCTATATATAATTACATGTAATTCTAACAGTAAATCGATCAATCTTCTTATCTTCATTATATAAAATTACGCTCCACTTCCATTTTCATTTTCAAGCTCGAAATTAATTGAATGTATTTGTCGAAAAGTATACAAAAAGAGAAAAAAAAA
**************************************************...(((((......(((.............((((((((......)))))))).............))).....))))).((((((.((..(((((((..(((.......))).))))))))).))))))......................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
V053 head |
M028 head |
SRR060670 9_testes_total |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................GGAACATTCAGGCGGTATGT......................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................TTTTCATTTACAAGCTCG.................................................. | 18 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................GGGCTGGTTCCCTAATACTA.................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................AAAAAATTGCGCTCCGCTTC...................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:3620642-3620892 - | dvi_13087 | GTCAACCCGAATGGATATCTGTGGCGAGGATTACGCTGCCTCTTCGCCTGGTAAGTCTGCCATATACTAAAAAGGGCTAGTTGGTCACGACCAAGTGATTAGGATATATATTAATGTACATTAAGATTGTCATTTAGCTAGTTAGAAGAATAGAAGTAATATATTTTAATGCGAGGTGAAGGTAAAAGTAAAAGTTCGAGCTTTAATTAACTTACATAAACAGCTTTTCATATGTTTTTCTCTTTTTTTTT |
| droMoj3 | scaffold_6473:8119871-8119905 - | CTTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATG-ATTTTAATGTA-TTTCCTTTCCTCTCTT | |
| droGri2 | scaffold_15203:10267657-10267687 + | TCTAACACCTAGGTGTATATATGACGAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAT | |
| droAna3 | scaffold_12929:1924648-1924681 + | AC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGCACAGCGTTCC-T---TTTTTTTATTTAATTTT | |
| droFic1 | scf7180000454045:861433-861451 - | TG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTTTTTTCCTTTTTT | |
| droEle1 | scf7180000491044:1127154-1127200 + | TA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCTACTTACAATAACA-ATTTTCATTTAATTTTGTATTTTTTCTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 05/16/2015 at 08:02 PM