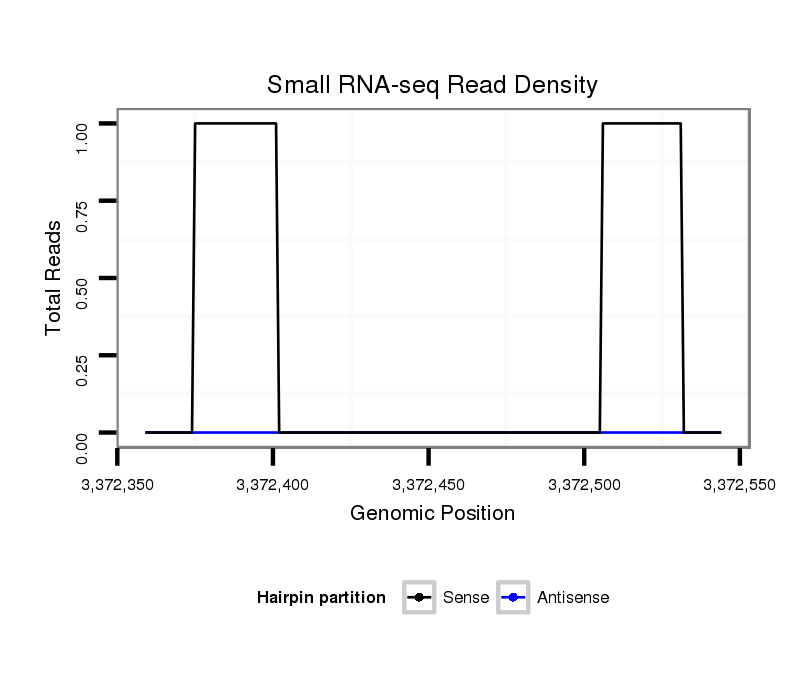
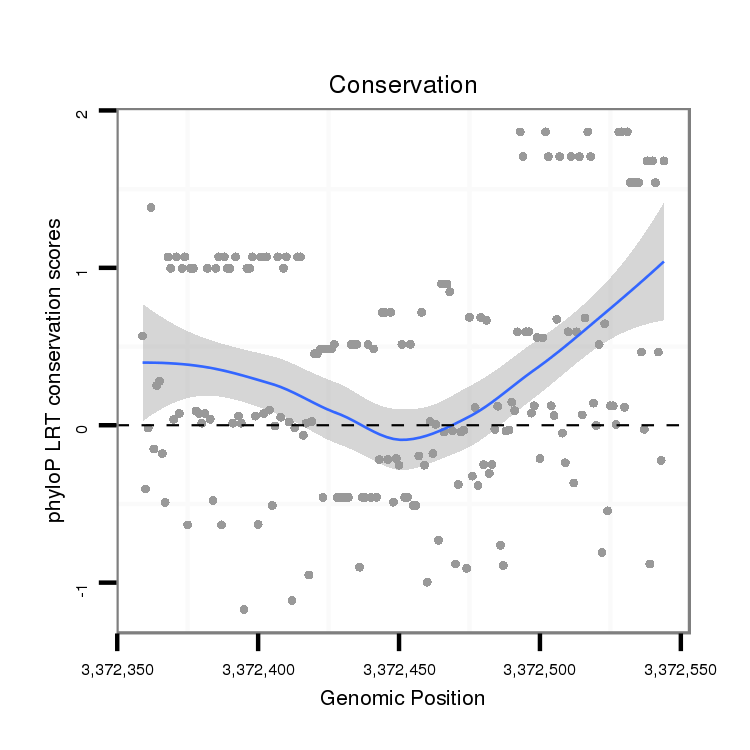
ID:dvi_13071 |
Coordinate:scaffold_12970:3372409-3372494 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_4086:3]; exon [dvir_GLEANR_4086:2]; CDS [Dvir\GJ19202-cds]; CDS [Dvir\GJ19202-cds]; intron [Dvir\GJ19202-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------################################################## CGCTGCATCTGCTGCATGGACGCGACCTATGGCACGCCCACCTGAAACATGTAGCTAGCCAGCAGCCCAGCTAGAATGGGATCTCATCTCTTATGATTGAAGTTCTATTGGTTTTCTATTTGGTTTTATTTTTCAGCCCAAGCACTTTGCACCACGCTACAACATCGCCAACTGGGGCATTTGCCT **************************************************......((((((......(((((..(((((((.(((((......)).))).))))))))))))......))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060669 160x9_females_carcasses_total |
SRR1106722 embryo_10-12h |
SRR1106726 embryo_14-16h |
V053 head |
M061 embryo |
SRR1106717 embryo_6-8h |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................TGGACGCGACCTATGGCACGCCCACCT............................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................TGCACCACGCTACAACATCGCCAACT............. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TGGTTTTCTATTTGGTTTGACT........................................................ | 22 | 2 | 2 | 1.00 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................CGGTTTTCTATTTGGTTTGATT........................................................ | 22 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................TGGTTTTCGATTTGGTTTGATT........................................................ | 22 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................TGGTTTTCGATTTGGTTTTACT........................................................ | 22 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................CATCGCATACTGGGGC........ | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
|
GCGACGTAGACGACGTACCTGCGCTGGATACCGTGCGGGTGGACTTTGTACATCGATCGGTCGTCGGGTCGATCTTACCCTAGAGTAGAGAATACTAACTTCAAGATAACCAAAAGATAAACCAAAATAAAAAGTCGGGTTCGTGAAACGTGGTGCGATGTTGTAGCGGTTGACCCCGTAAACGGA
**************************************************......((((((......(((((..(((((((.(((((......)).))).))))))))))))......))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060671 9x160_males_carcasses_total |
V053 head |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................GTCGGGTACGTGAATCGTGC................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| .........................................................TGCTCGTAGGGTCGATCTTA............................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 |
| .................................................................................GGAGTAGAGAATCCTAGCT...................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ..................................................................................GAGTAGAGAATCCTAGCTG..................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:3372359-3372544 + | dvi_13071 | CGCTGCATCTGCTGCATGGACGCGACCTATGGCACGCCCACCTGAAACATGTAGCTAGCCA--GCAGCCCAGCTAGAATGGGATCTC-----ATCTCTTATGATTGAAGTTCTATTGG---------TTTTCTATT---------TGGTT----------TTATTTTTCAGCCCAAGCACTTTGCACCACGCTACAACATCGCCAACTGGGGCATTTGCCT |
| droMoj3 | scaffold_6473:7825558-7825729 + | CGCTGCATCTGCTGCACGGACGCGGCCTGTGGCATGGCCATTTGAAGAACGTACGTAGCTATAGCA--------------------T-----ATTTCTC------GAGTTTCCATTGGAGTCATTGACATTT-GCTCTGGCTCTT-----------------CCCCCTCAGCCCAAGCACTTTGCGCCGCGCTACAACGTCGCCAACTGGGGCATCTGCCT | |
| droGri2 | scaffold_15203:10501886-10502052 - | CGCTACACTTGTTGCACGGACGCGGACTGTGGCACGGCCACTTGAAAAATGTGAGTAGCAT-----ACCCAATCGAAATCAAACCCCAGGGGATCTGCTACAAAAAAAAACA-------------------------------------------------CATTTTGCAGCCCAAGCACTTTGTGCCACGTTACAACATCGCCAACTGGGGTATCTGTGT | |
| droWil2 | scf2_1100000004963:2870122-2870276 + | CATTGCACATGCTACAAGGTTTTGGTCTTTGGAACTCCCACATAAGTAATGTAAGTACAAA------------------------------------------------TCCAAATGA---------GTACGTGTG---------TAGACTTATATCCATCCAATCATCAGCCAAAGCACTTTGTGCCACGATACACCGTGGCGAATTGGGGCGTGTGCAT | |
| droAna3 | scaffold_13117:1263994-1264049 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCAGCCCGGACACCGTGTGCCCCGCTACTCCCTGATCAACTGGGGCATCTGCGT | |
| droBip1 | scf7180000396425:850523-850596 - | TCC----------------------------------------------------------------------------------------------------------------------------------ATCTTGCCTTTT-----------------TCATTTCAGCCCGGACACCGTGTACCCCGCTACTCCCTGATCAACTGGGGCATCTGCGT | |
| droKik1 | scf7180000299277:473094-473161 + | TTTTATT---------------------------------------------------------------------------------------------------------------------------------------------------------CCCATTTGCAGCCCGGTCACCGAGTGCCCCGCTACTCGCTAATCAACTGGGGCCTGTGCGT | |
| droTak1 | scf7180000415184:120998-121040 - | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTAGTTTAGCAACTTTGCATCATGCAACAACAGCGCAAATT------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/19/2015 at 05:51 PM