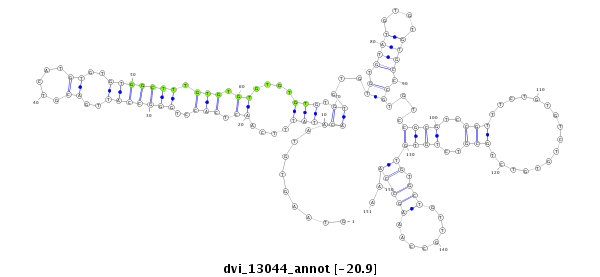
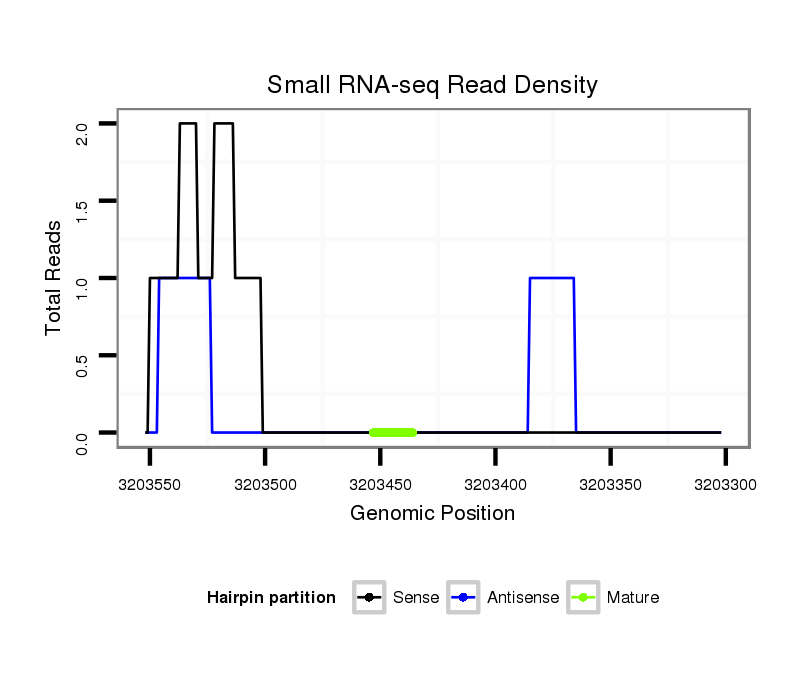

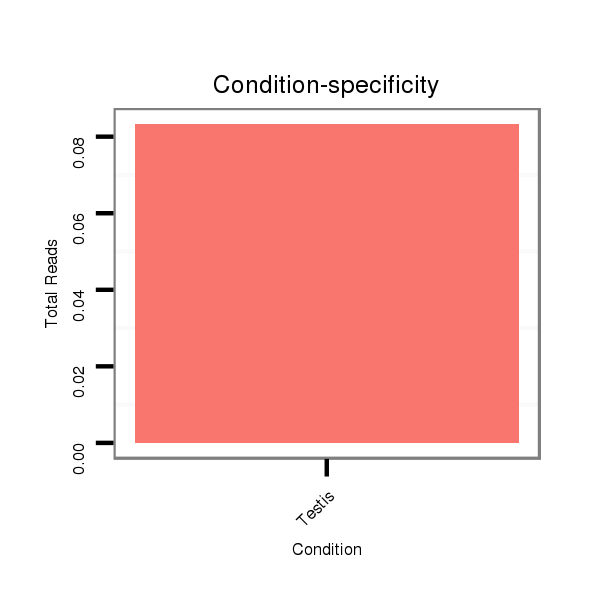
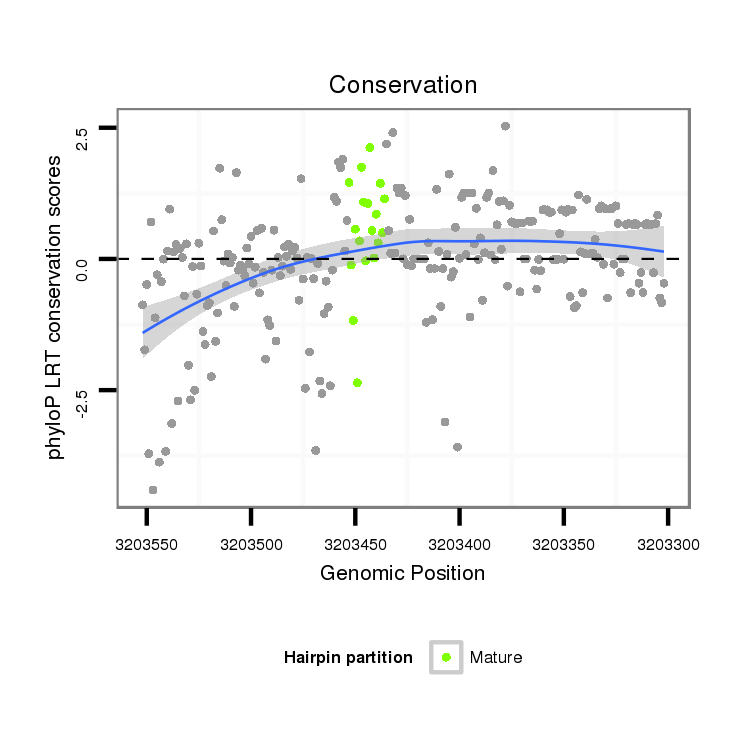
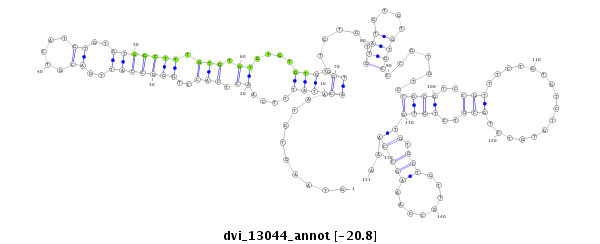
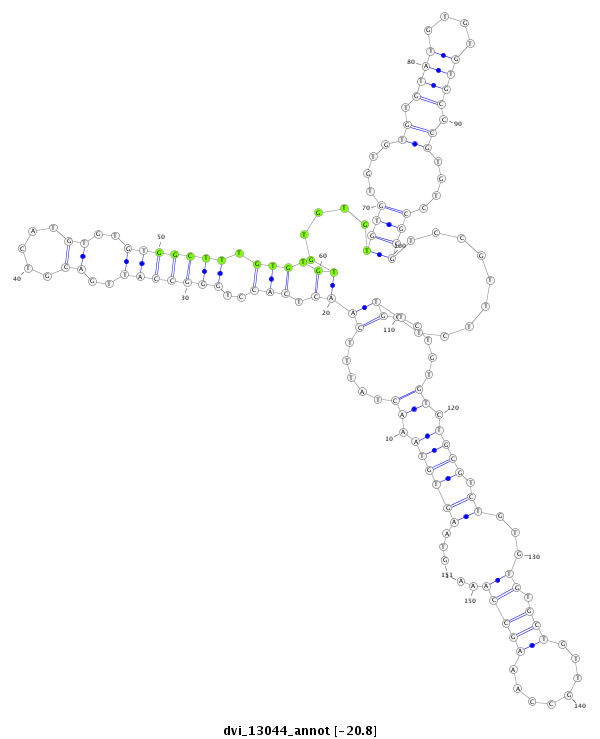
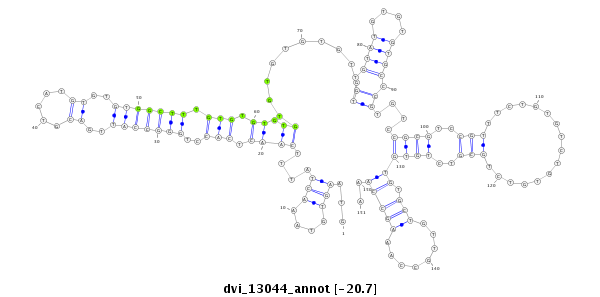
ID:dvi_13044 |
Coordinate:scaffold_12970:3203352-3203502 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.8 | -20.8 | -20.7 |
 |
 |
 |
exon [dvir_GLEANR_3793:3]; CDS [Dvir\GJ18980-cds]; intron [Dvir\GJ18980-in]
| Name | Class | Family | Strand |
| (CA)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CTTTACCAGCAACATCATCATCAGCAGCTGTTGTCGAGCAGCTTCATGCGGTAAGTGTAAACTATTTCAACTCACCTGGGCCATTGACGTCATGTGTGTGGCTTTGTGTGTGTGTGTGTGTGTGTGTGTATGTGTGTGCCCGTGTCCGCGTCCGTTTCTGTGTCTGTGTCTGCGTCTGTGTGTGCTGTTGCCAAAGCCAAAAACTCGGGATGTGGTTGCCAGCGGGGATTGGGTGGACGAGCAGCGACAGA **************************************************..........(((((....((.(((..(((((((..((.....))..))))))).))).))....))).))...((.((((....)))).))....((((..(((................)))..))))((.(((........)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060667 160_females_carcasses_total |
SRR060678 9x140_testes_total |
V053 head |
M028 head |
M047 female body |
V047 embryo |
V116 male body |
SRR060672 9x160_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060680 9xArg_testes_total |
SRR060669 160x9_females_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
M061 embryo |
SRR060666 160_males_carcasses_total |
SRR060673 9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................TTGTCGAGCAGCTTCATGCGG........................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............CATCATCAGCAGCTGTTGTCGAGC.................................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TTACCAGCAACATCATCATCA.................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................AGCTTAAGCCGGTAAGTGTA................................................................................................................................................................................................ | 20 | 3 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................GCAGCTGTTGTCGAGCAA.................................................................................................................................................................................................................. | 18 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TCAAAAACTCGTGATGTTGTTG................................. | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........GCAACATCATCATCAGCAGC............................................................................................................................................................................................................................... | 20 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................GAATGGTTGGACGAGCACC...... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CAACATCATCATCAGCAGC............................................................................................................................................................................................................................... | 19 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GGCTTTGTGTGTGTGTGT...................................................................................................................................... | 18 | 0 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................AGCTGGGATTGAGTGGACGT........... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GGTGTATGAGTGGGCCCGTG........................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................CCGTTACTTTGTCGGTGTCT................................................................................ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................CATCAGTTTCTGTGTCTGTGTA................................................................................. | 22 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................TGGCTTTGTGTGTATGTG....................................................................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................CGTGTGTGCCCGTGTCCGTGT.................................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................AACCTTAGCCATTGACGT................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GAAATGGTCGTTGTAGTAGTAGTCGTCGACAACAGCTCGTCGAAGTACGCCATTCACATTTGATAAAGTTGAGTGGACCCGGTAACTGCAGTACACACACCGAAACACACACACACACACACACACACATACACACACGGGCACAGGCGCAGGCAAAGACACAGACACAGACGCAGACACACACGACAACGGTTTCGGTTTTTGAGCCCTACACCAACGGTCGCCCCTAACCCACCTGCTCGTCGCTGTCT
**************************************************..........(((((....((.(((..(((((((..((.....))..))))))).))).))....))).))...((.((((....)))).))....((((..(((................)))..))))((.(((........)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
M028 head |
V047 embryo |
V053 head |
M027 male body |
M061 embryo |
SRR060661 160x9_0-2h_embryos_total |
SRR060680 9xArg_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060659 Argentina_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................TGGACCGGATAACTGCACTAC............................................................................................................................................................. | 21 | 3 | 2 | 30.50 | 61 | 22 | 14 | 12 | 3 | 3 | 3 | 1 | 2 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................TGGACCGGATAACTGCACTA.............................................................................................................................................................. | 20 | 3 | 3 | 13.33 | 40 | 17 | 10 | 4 | 2 | 4 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................GTGTGGACCGGATAACTGCA................................................................................................................................................................. | 20 | 3 | 3 | 8.00 | 24 | 6 | 6 | 0 | 10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................GTGGACCGGATAACTGCACTA.............................................................................................................................................................. | 21 | 3 | 2 | 6.00 | 12 | 4 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ......................................................................GTGTGGACCGGATAACTGC.................................................................................................................................................................. | 19 | 3 | 12 | 3.33 | 40 | 11 | 11 | 0 | 10 | 6 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GGACCGGATAACTGCACTAC............................................................................................................................................................. | 20 | 3 | 5 | 3.00 | 15 | 8 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................GTGGACCGGATAACTGCACTAC............................................................................................................................................................. | 22 | 3 | 2 | 2.00 | 4 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GGACCGGATAACTGCACTA.............................................................................................................................................................. | 19 | 3 | 11 | 1.55 | 17 | 7 | 6 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................GTGGACCGGATAACTGCA................................................................................................................................................................. | 18 | 2 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GTCGTTGTAGTAGTAGTCGTCGA.............................................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................CAGACGCAGACACACACGAC................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TGGACCGGATAACTGCACT............................................................................................................................................................... | 19 | 3 | 10 | 0.80 | 8 | 5 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................AACCCACCCGCTCGTCGCTG... | 20 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................GTGGACCGGATAACTGCACT............................................................................................................................................................... | 20 | 3 | 5 | 0.40 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CACAGACGCAGACACACAC................................................................... | 19 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................CACTCACGGGCACAGGCT...................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................GCGTGGACCGGATAACTGC.................................................................................................................................................................. | 19 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................TGGGGACCCGATAACTGCA................................................................................................................................................................. | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................GTGGACCAGATAACTGCAC................................................................................................................................................................ | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....TGGTCGTTATAGTAATAG..................................................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:3203302-3203552 - | dvi_13044 | C-TTTACCAGCAACATC-ATCATCAGCA-----GCTGTT-----------GTCGA---GCAGCTTCATGC----G------------------------------------GT-AAGTGTAAACT-ATTTC------------A----------------------------ACTCACCTGGGCC---ATTGAC-------------GTCA----------------------------------------------------------TGTG--T------GTGGCTT--TGTGTGTG----------------T--GTG------------------------TGTGTG------------------TGTGTGTATGTGTGTGCCCGTGTCCGCGT-----------------------------------------CCGTTTCTGTGTCTGTGTCTGC-------------------G----TCTG------TGTGTGCTGTTGCCAAAGCCAAAAAC--------------------------------------------------------------------------------------------------------------T-------------------------------------------------------------------------------------------------------------------------CGGG--ATGTGGT--TGCCAGCGGGGATTGGGTGGACGAGCAGCGACAGA---------------------------- |
| droMoj3 | scaffold_6473:4063198-4063438 - | C-ATCAACAGCAACATC-ATCATCAGCA-----GCTGCT-----------GTCGA---GCAGCTTTATGC----G------------------------------------GT-AAGTGTAAACT-ATTTC------------A----------------------------ACTCACCTGGGCC---ATTGAC-------------GTCAGCGTGTTC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCAAAGCCAAAAACCGGGCATTTCAAGTGTGTGTGTGTGTGTGTGTGGAGGGGGGAGGGGGGCAATCCCCCT--------------------------------------------------------------------------CGGTACAGCATGTGAGTGCGAGGAGCTGGGGGATTCGAGCTTGGGGTG----GGATACCGGGCCATGGG-----------------------------------------------------------------------------CGGA---------------------------- | |
| droGri2 | scaffold_15081:3942390-3942604 + | C-ATTAATATCAACAAGCAACAGTGGCG-----C----------------------------------------C----------GAAAAGTATGTGTTAC-----------------ATTTGTA-AGTAGGAGGACACATGT------------------------------GT---ATATGAGTGTAGTTATGT-------------AAATGTG-------------------------------------------------------TG--T------GTGTGTG--TGTGTGTG----------------T--GTG------------------------TGTGTGTGTGTGTGTGT------------GTG------TGTGTGTGTGTGTGTGTG--TG---------------------------T------GT------GTGTG------TGT-------------------G----T--------------------------------------------------------------------------------------------------------GTGTGTGTGT--GTGTGTGTGAGCAAGCAAAAGGCAGCAAGT-------------------------------------------------------------------------------------------------------------------------T-----------------------------------------------GGG--------------------------- | |
| droWil2 | scf2_1100000004822:717218-717373 - | A-TCAACACAGGAT-----------GTTTTTGCGCTCA--------------------------TTATGC----G------------------------------------GC-AGCTGTATT-------C------------A----------------------------C--------------CACGTGTGT----------------------------------------------------------------------------GCGTGT----GT----GT--GTGTGTG--------TG--------------------TGTGTGT------GTGTGTGTGT----------------------GTG----------------------TG--TGTGTGTGT------GTG-----------TGTGTGTGTGTGTGTGTGTCTGTGTCTGT-------------------G----T------CTATGTCT-------------------------------------------------------------------------------------------------------TTT--GT--------------------------------------------------------------------------------------------------------------TG--------------------------------------------------------------------------------C---------------------------- | |
| dp5 | XL_group1a:2069610-2069795 + | C-TATATCAATGCT-----------TTCT-----------------------------------------------------------------------------------------------------------------------------------------------------CTGTGTC---T---------------------------------------------------------------------------------------------------------------CTGACTG------TGTGT--GTGTGTGTGTGT----------GTGTGTGTGTG----------------TGTGTGTG------------------------TGT--GTGTGGTGGTGTGTTGTT--------T------GT------TTGTG------CGC-------------------G----T--------------------------------------------------------------------------------------------------------GTTTATATTT--GTGT--GG--------------------------------------------------------------------------------------------------------------TG-------------CTCCTTCCGGTGAAAAATTGACATATTTGGTGCTGCCAACTGGAATC--GTCGACTGCCAGCAACTC-G--------------------------- | |
| droPer2 | scaffold_4:6832494-6832683 - | T-TTA-CCATT----------TGCCATG-----AT----GAGCAGCCAGCAGCCAGCAGCAGTTGCAG-----GGA------------------------------------G-AAATGTAGGAA-AGCTT------------A----------------------------ACCCTACTCGCAC---ATTCAT-------------TCGAG----------------------AACGCACACAGGGG-TCCC--------CC-------------CT--------------GTCTGTG----------------T--GTG--TG----TGT------------------G------TGTGT------------GTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTGTGTGTGTATCCACGTTGTTTGTTTGTTTGTTTGTTT--GTTT--G-------------------------------------------------------------------------------------------------------------TTTG------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13334:1360048-1360198 - | T-GTG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG--T------GTGTATG--TGTGTGTG----------------T--------------------------GTGTGTGTGTGTGTGTGTGT------------G----------------------------T--GTGTGTTGGTGTGTTAGCTGG-TGTGT------C----------------TATGT-------------------GTTAGT--------------------------------------------------------------------------------------------------------GTGTGTGTGT--GTGG--GTTA----------------------------------------CCTGTAGAAGA--------------------------------------CCGT----------------TAACTAAGCACACG--------------------GC--AAATGTT--TGCCAGCCGGGA--------------------------------------------------- | |
| droBip1 | scf7180000396543:156401-156520 - | C-CTCAGTCGCAGCACC-AGCACCAGCA-----GCCGTC-----------AG--T---GCTG-----------ATA-----------AA------------------------------------------------------------------------------------------------AACGTACGTAT-------------ATGT---------------------------------------------------------------GTGTGT------------------GTGACTCAG--------------------------------------------------------------------------------TCTCTGTATGTA--TG---------------------------T------AT------GTATG------TGT-------------------G----T--------------------------------------------------------------------------------------------------------GTATGCGTGT--GTGA--G-------------------------------------------------------------------------------------------------------------CTTC------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302808:901417-901587 - | G-TTCGACGCTGCCAGC-GCCGACTGTT-----GCTCAT-----------CTCC-----CGAC-----------G---------------------------------------------------------------------------------------------------------ACGCC---ATTGT-------------------C-------------------------------------------------------CTGTG--T------GTGCTCT--GGTATGCG----------------T--------------------------GTGCGCGTGCATGTGTCCGT------------G----------------------------T--CCATGT---------------------------CCGTGTCTGTGAGTGTATCTGT-------------------G----TCTGTGGCGG-------TGTCCTGAAAGCTGG-AGC--------------------------------------------------------------------------------------------------------------T-------------------------------------------------------------------------------------------------------------------------GGCA--ATGCGGC--CGG------------------------------TAG--------------------------- | |
| droFic1 | scf7180000451789:487753-487957 - | C-AACAGCAGCAACAAC-AGCAGCAGCA-----GCAGCA-----------AC--A---GCAGC-----------AACAACAGCAAC--------------------------------ATCAGAA-AGAGCA-GC--------A----------------------------A-----------CATCAGCGGC-------------GGCAACGCGTTC------AA--GTGAAAA-GTCCGCAGCTGCTGCCCGCACT--TCATGTAC----------------------------T--ACGATTC------CAAAT--GTGTGAGTGTGC----------CAGTGTGTGTG----------------TGTCTC------------------------TATCT--GTGTGC---------------------------------------A------AGT-------------------G----T--------------------------------------------------------------------------------------------------------GTGAGT--GTGCGAGT--GT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000490214:839958-840163 - | C-AGTAGCAGCAGCATC-ATAAGCCCTG-----AA------TCCGCCGGCGTCTGACCGCAAATTC---------GTAACA------------------------------------------------TC------------A----------------------------ACAAA------------------AATGAAATTAACATCAAA---GTTGCCCAACCAG--------------------------CCAT-------------GCATGT----AT----CTG--TGTGTG--TG--------------------------TGTAAGTGTGTGTGTGTGTATGTGTATGTGTTT------------G----------------------------T--GTGTAT---------------------------CGGTGTGTGTGTGCGTATCTGT-------------------GTT-----------------------------------------------------------------------------------------------------------------GTAT--ATTT--GT--------------------------------------------------------------------------------------------------AG-----CCATGTG------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000777516:2970-3146 + | C-ATCAGCAGCAACAGC-AGCAGCAACA-----ACAACA-----------GCCA-----CACC-----------AACAACAGCAACAAAAGCGCACTTCAA-----------------------------------------------------ACGCCG-------------------------------CA------------------------------------------------------------------------------TG--T------TCGTATCGG--TGTGTTGC------TG--------------------CCC--CTGTGTGTG--TGTGTT----TTAATGG------------AAG------AGAGAGGACATGCAGCTG--TA---------------------------A------AT------ATCCC------TCA-------------------G----T--------------------------------------------------------------------------------------------------------ATGTGTGTGA--GTGT--G-------------------------------------------------------------------------------------------------------------TGTG------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000301760:297547-297675 - | T-TTCAGTTTAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTGTTCGATGGCGT-------------GGGCGTG-------------------------------------------------------TG--T------GTGTGTG--TGTGTGTG----------------T--GTG------------------------TGTGTGTGTGTGTGTGT------------GTG------TGTGTGTGTATGTGTGTG--TG---------------------------T------GT------GTGTG------TGT-------------------G----T--------------------------------------------------------------------------------------------------------GTGTGTGTGT--GTGT--GT--------------------------------------------------------------------------------------------------------------TG------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415381:115313-115437 - | T-GTG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG--T------GTGTGTG--TGTGTGTG----------------T--GTG------------------------TGTGTGTGTGTGTGTGT------------GTG------TGTGTGTGTGTGTGTGTG--TG---------------------------T------GT------GTGTG------TGT-------------------G----T--------------------------------------------------------------------------------------------------------GTGTGTGTGT--GTGT--GTTGGC------------------CAGGATAAGTATCCACTAAG----------------------------------------------------------------CAGTTG------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409182:66453-66642 + | ACATCATCATTAGCAT--------------------ATT-----------GACAT---GTGGCCAAATTT----G------------------------------------GTCACGTTCCATTTCATTTC------------AATTCATTCTGAC--------------------------------ATTGCC-AACGGAGTTCGAATCAAT---TTCCCATCTCCAG--------------------------CCAAGTTCG-----TGT----------------ATG--TGTGTTT--------------------G--------TGT------------------G------TTTGTGTGTGTGTGT--G--------------TGTGTGTGTGTG--T--GTGTGT---------------------------------------T------TGT-------------------G----TGT------------------------------------------------------------------------------------------------------GT--GT------GTGT--G-------------------------------------------------------------------------------------------------------------TGTG------------------------------------------------------------------------------------------------------------- | |
| dm3 | chrX:4358968-4359157 + | C-AGCAGCAGCAGCAGA-AGCGGCAGCA-----GCTCAG-----------CATCA---GCATCATTAGAGCCAGAT---------T--A------------------------------------------------------------------------------------------------TACACTCATCT-------------GAAT---------------------------------------------------------------CTGTTT------------------ATGTTTCAGTATCTGT--------------------------ATCTGTATCTGTATCTTTGT------------CTC------TGTCTGCGCCT----------------------CTGCTTTGTAACCGTGG------C----------------TTTGTCTTTAGACTTTGT------G----T--------------------------------------------------------------------------------------------------------GCGTGT--GCTCGAGT--GTCGCC-----------------------------------------------------------------------------------------------------------------------------------------------------------A-------------------------------AAG--------------------------- | |
| droSim2 | 2l:7720902-7721121 - | C-GATAATAGCAACAAC-AACAGCGACA-----C----------------------------------------A---------ATAAAAGAATTTGTCAGGCGGCGTTTGTTTTTGTTCACCCTCTCTCT------------TTCTCTCTCGCCC--------------------------------ATCGTC-T------------GTGTCGTGCTT------TCCG--------------------------CTCCGTCTC-----TGT----------------CTG--CCGGCCT-TCGTC--------------------------------------------------GCGTGT------------GTG------CGTGTGTCCCCGTGTGCG--TG---------------------------C------GT------GCTTG------TGT-------------------G----T--------------------------------------------------------------------------------------------------------GTGTGTGTGC--GTCC--GTCG----------------------------------------TCTGTGCAGCGCAAAGAT-----------------------------------------------A---------------------------------------------------A-------------------------------AAA--------------------------- | |
| droSec2 | scaffold_0:2225759-2225908 - | C-ATGTTGGCCATTG-------------------------------------TC-----C-----------------------------------------------------------------------------------------------------------------------------TGCGGTGGTGT-------------GTGT---------------------------------------------------------------GT--------GTG--------T--GTGTGTGTGTGTGTGT--------------------------G--------------TGTGT------------GTG------TGTGTGTGTGTGTGTGTG--TG---------------------------T------GT------GTGTG------TGT-------------------G----T--------------------------------------------------------------------------------------------------------GTGTGTGTGT--GTGT--GT--------------------------------------------------------------------------------------------------------------TG-------------CTCCC-------------T-----------------------------------------------GGGCACAATGGTCAGCAAAATGCGGTCTCC | |
| droYak3 | X:13119023-13119212 - | C-ATCATCATCATCATC-GGCATCGACA-----ACTCAG-----------GTCAG---G----------------------------------------------------------CCCCGCTTCTGTCC------------GTCTCTTTCTAACGTCGTATGTCTGTCTCTCTCACTTGCATC---CTTGAC--------------------------------------------------------------------------------------------------------------CTG------CGTGT--GTGTGTGTGTGT----------GTGTGTGTGTG----------------TGTGTGTA------------------------TGT--GCGTTCTGACATCCTGAAAGA--------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGTGTTT--GTGT--TT-----------------------------------------------------------------------------------------------TGTGA-----CAGCGTC------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4690:1734989-1735149 + | C-ATCAGAATCAGCATC-ATTAGAGCCG-----G----------------------------------------AT---------T--A------------------------------------------------------------------------------------------------TACACTCATCT-------------GAAT---------------------------------------------------------------CTGTTT------------------ATGTTTCTGTTTCTGT--ATC------------------------TGCATCTGTATCTGTAT------------CTG------CATCTGTATCTGCAT-------------------------------GTGG------C----------------TTTGTCTTAAGATTTTGTGTGCGTC----T--------------------------------------------------------------------------------------------------------GTGAGT--GCTCGAGT--GTCGCC-----------------------------------------------------------------------------------------------------------------------------------------------------------A-------------------------------AAG--------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/19/2015 at 05:59 PM