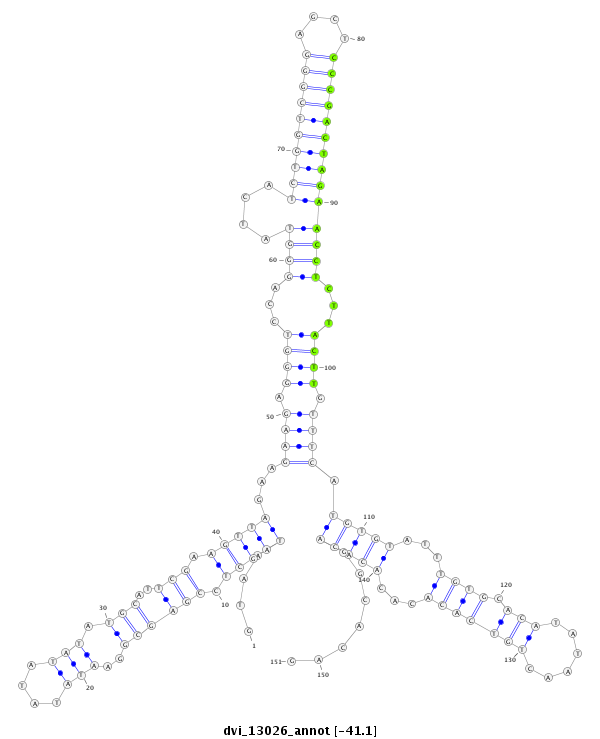
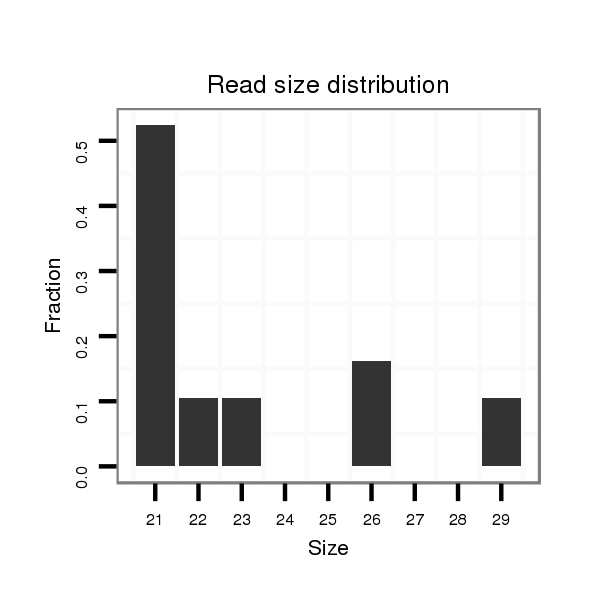
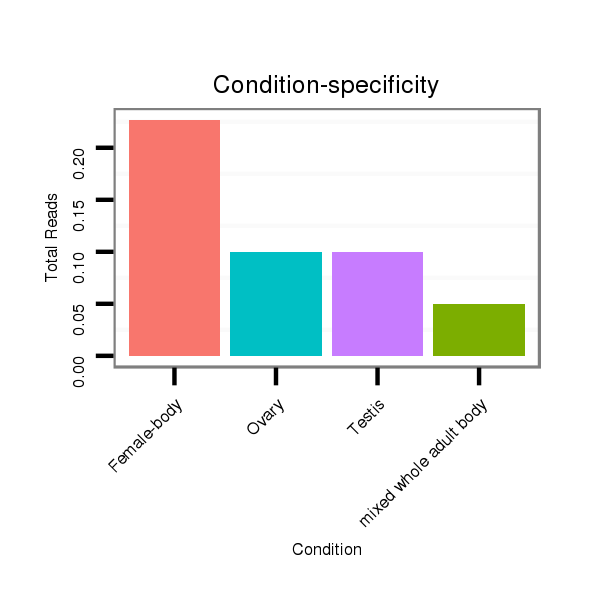
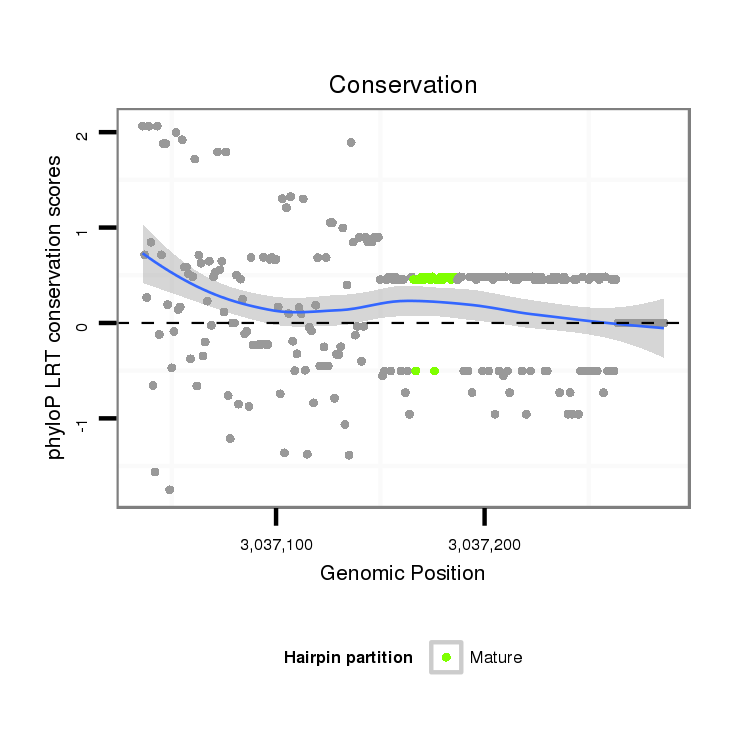
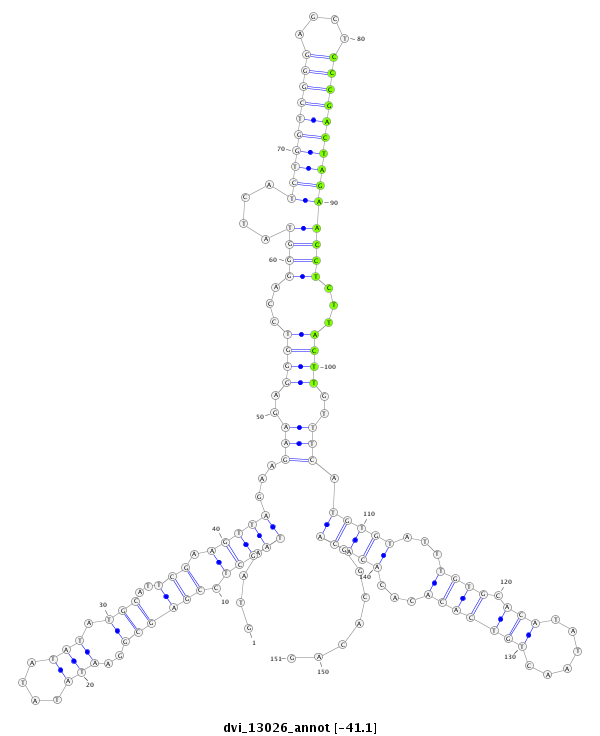
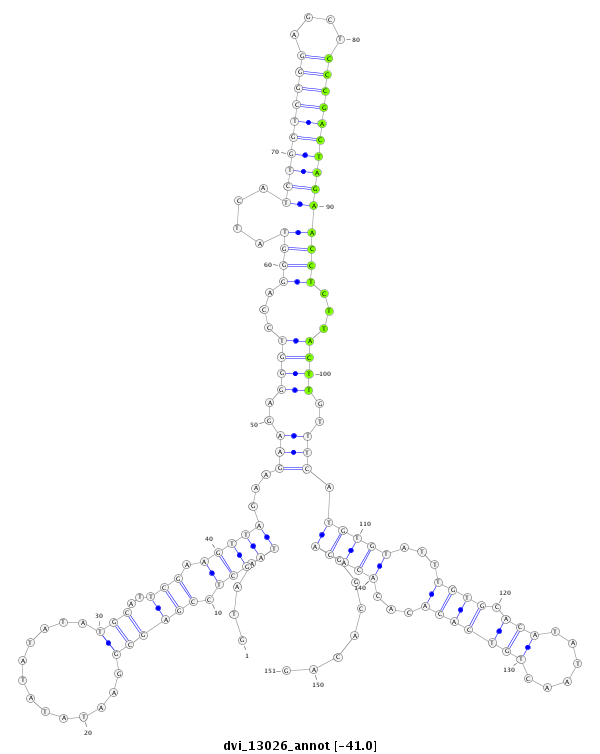
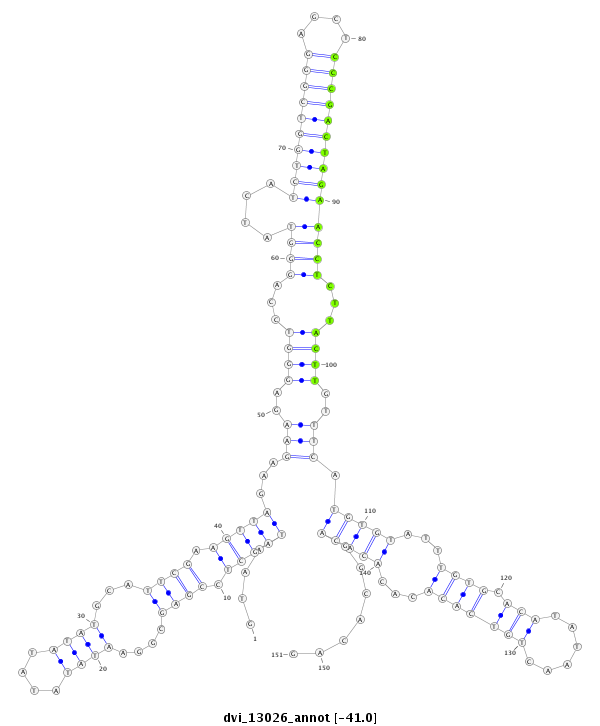
ID:dvi_13026 |
Coordinate:scaffold_12970:3037086-3037236 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -41.1 | -41.0 | -41.0 |
 |
 |
 |
CDS [Dvir\GJ19186-cds]; exon [dvir_GLEANR_4070:1]; intron [Dvir\GJ19186-in]
| Name | Class | Family | Strand |
| Helitron-1N1_DVir | RC | Helitron | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACATAACTCCGCGAGGAGATATGACGTTCCAAAACGACATAATATCTCGGGTAATAGCTCCGAGCGGAATATATATATATGCATTCGAAGTTAGAAGAAGAGGGTCCAGGGTATCATCTGGTCGGGAGCTCCCGACTAGAACCTCTTACTTGTTTCATGTGTATTTGTGCACATATAACTGTCACACACACACAGGCACAGCTGGAGCCCTACACTGAACTGTGCGGCACACAATTTTTATTTAATAAATA **************************************************....(((((.((((((..(((....))).)))..))).)))))...((((.((((...((((....((((((((((....))))))))))))))...)))).)))).(((((...((((.(((......)))))))...))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
M047 female body |
GSM1528803 follicle cells |
SRR060658 140_ovaries_total |
SRR060678 9x140_testes_total |
SRR060657 140_testes_total |
SRR060665 9_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................ATAATATCTCGGGTAATAGCT................................................................................................................................................................................................ | 21 | 0 | 20 | 0.60 | 12 | 11 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AGGGTCCAGGGCATCGTGTG................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CGGGTAATAGCTCCGAGCGGA....................................................................................................................................................................................... | 21 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAGAAGAAGAGGGTTCAGGGTATCA....................................................................................................................................... | 25 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ..................................................................................................................................CCCGACTAGAACCTCTTACTT.................................................................................................... | 21 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTAGAACCTCTTACTTGTTTC............................................................................................... | 21 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................ATATCTCGGGTAATAGCTCC.............................................................................................................................................................................................. | 20 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................CCGACTAGAACCTCTTACTTGTTTCA.............................................................................................. | 26 | 0 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................AAGTTAAAAGGAGAGGGT.................................................................................................................................................. | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TCGGGAGCTCCCGACTAGAACCTCTTACT..................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................GAGCTCCCGACTAGAACCTCT......................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................CCCGACTAGAACCTCTTACTTGT.................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CGGTTAATAGCTCCGAGCGGA....................................................................................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................CGGGAGCTCCCGACTAGAACCT........................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................ATAATGTCTCGGGTAATAGCT................................................................................................................................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................CCGACTAGAACCTCTTACTTGATTC............................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TGTATTGAGGCGCTCCTCTATACTGCAAGGTTTTGCTGTATTATAGAGCCCATTATCGAGGCTCGCCTTATATATATATACGTAAGCTTCAATCTTCTTCTCCCAGGTCCCATAGTAGACCAGCCCTCGAGGGCTGATCTTGGAGAATGAACAAAGTACACATAAACACGTGTATATTGACAGTGTGTGTGTGTCCGTGTCGACCTCGGGATGTGACTTGACACGCCGTGTGTTAAAAATAAATTATTTAT
**************************************************....(((((.((((((..(((....))).)))..))).)))))...((((.((((...((((....((((((((((....))))))))))))))...)))).)))).(((((...((((.(((......)))))))...))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060684 140x9_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
M047 female body |
SRR060678 9x140_testes_total |
SRR060687 9_0-2h_embryos_total |
V053 head |
SRR060656 9x160_ovaries_total |
SRR060683 160_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................ACCATCGGGGCTCGCCTTA..................................................................................................................................................................................... | 19 | 3 | 12 | 0.25 | 3 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................CCGGAGCTTCAATCTTCTTC....................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TATTGAGGCGCTCCTCTATACT................................................................................................................................................................................................................................... | 22 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................GGGCTGATCTTGGAGAATGAACAAAG............................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................ATTGACAGTGTGCGTGTC.......................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................................................TAAAAATAGATTATTTAT | 18 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............TCCTCTATACTGCAAGGTTTTGC....................................................................................................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................CGAGGGCTGATCTTGGAGAAT....................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:3037036-3037286 + | dvi_13026 | -ACATAACTCCGCGAG-G------------------------AGATATGAC-------------------------GTTCCAAAACGACATAATATCTCGGGTAATAGCTCCGAGCGGAA-TATATATA--------------------------T------------ATGCATTCGAAGTTAGA--AGAA-GAGGGTCCAGGGTATCATCTGGTCGGGAGCTCCCGACTAGAACC-TCTTACTTGTTTCATGTGTA-TTTGTGCA---------CATATAACTGTCACACACACACAGG-CACAGCTGGAGCCCTACACTGAACTGTGCGGCACACAATTTTTATTTAATAAATA |
| droMoj3 | scaffold_6680:22241116-22241302 - | TATGTATGTGTGCATA-A------------------------AAATGCAGA---------AT--------------GATTAGA--------------------------------------------------------------------------------ATAT---------------CAGCT--TA-GAGGGTTCAGGGTATCTCCTAGTCGAGCAGTCTCGACTAGAGCCTTCTTACTTGTTCTGTTTGTAACTTATGGATTCTACTTGTAAACCACTGTTAGATACACACGAGCCACATCTGCCCCCGCATGCTAAGCTTTATGAC----------------------- | |
| droWil2 | scf2_1100000004515:1741992-1742114 + | -ACATATCTCCGCGCG-A------------------------AATTATGTC-------------------------GTTTCAGAACGACAAATA--TTCCTAA----------------------------TCTAA--ATAACTAAACAACAGAATCGTTGAAATATATTACAATATGATTTAAACCAGGA-AAGTATTTAGGGTAT----------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | Unknown_singleton_1421:182-268 + | -ACATATCTCCGCGCG-G------------------------AGCTATGAC-------------------------GCTTAGAAGCGACATATC--TCCGCGCAGCGATTTTGAGCAAAT-TATGCCGA--------------------------T------------ACACCA---------------TCAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droPer2 | scaffold_6336:1397-1483 + | -ACATATCTCCGCGCG-G------------------------AGATATGAC-------------------------GCTTAGAAGCGACATATC--TCCGCGCAGCGATTTTGAGCAAAT-TATGCCGA--------------------------T------------ACACCA---------------TCAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droKik1 | scf7180000300620:32-136 + | -ACATAACTCCGCGCG-G------------------------AGATATGTCGTTTGTAGTTCGGCGGAGATATGTCGCTTTAAAGCGACATAAC--TCCGCGC-------------GGAGATATGTCGTT--------------------------------------TGTAG-------TTCGG--CGG-AGA------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEle1 | scf7180000489519:6911-6971 - | -ACCTAATTTCGCGGG-G-AAAAACGACATAATTTCAC-CG-----------------------------------GCCTCGAAACTATAAATA--TAT--------------------------------------------------------------------------------------------ATA------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droRho1 | scf7180000778200:9123-9184 + | -ACATATCTCCGCCCTTCTGCAAACGACATATCTCCGCGCGGAGAT----------------------------------------------------------------------------GTGACGTT--------------------------------------AAAAA-----------------AAGA------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droBia1 | scf7180000299101:14197-14252 + | -ACATATTTCCGCGCG-G------------------------AGATATGAC-------------------------GTTTCAAGACGACATATC--TCCGCGC-------------GGAG------------------------------------------------------------------------TA------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droTak1 | scf7180000412279:11805-11893 + | -ACATAAGTTCGCGGCAG------------------------A--------------------------------------AAAACGACTTAAT--TTCGCCG-------------GCACATGTATAGTTATATATATATATATGTAAAACATAGT------------TCACGA---------------TATCA------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droYak3 | v2_chr2L_random_001:396002-396021 - | -ACATAACTCCGCGAG-G------------------------AGAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4690:18024903-18024943 - | -ACATTATTCCGCGCA-G------------------------AGATATGAC-------------------------GTTTCATA-GGACATAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/19/2015 at 05:57 PM