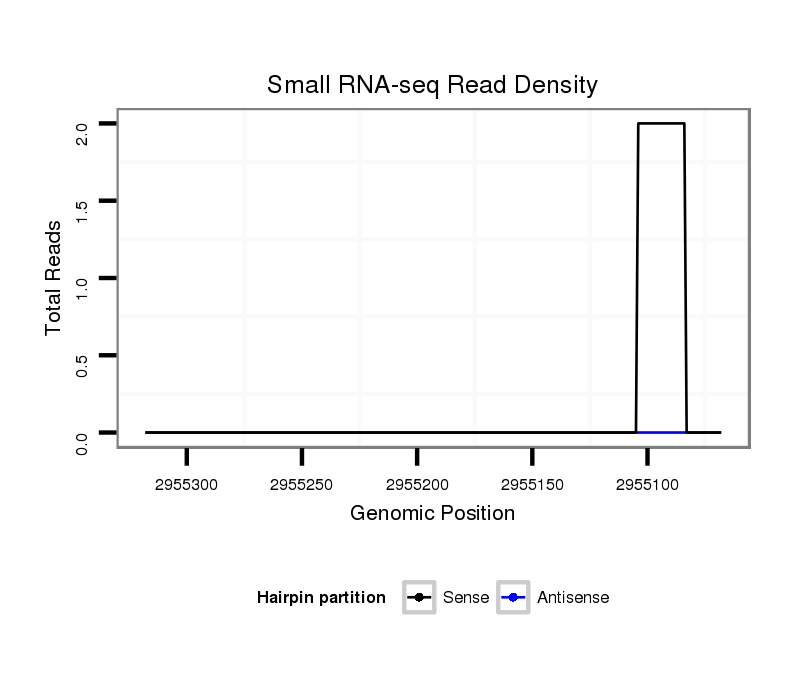
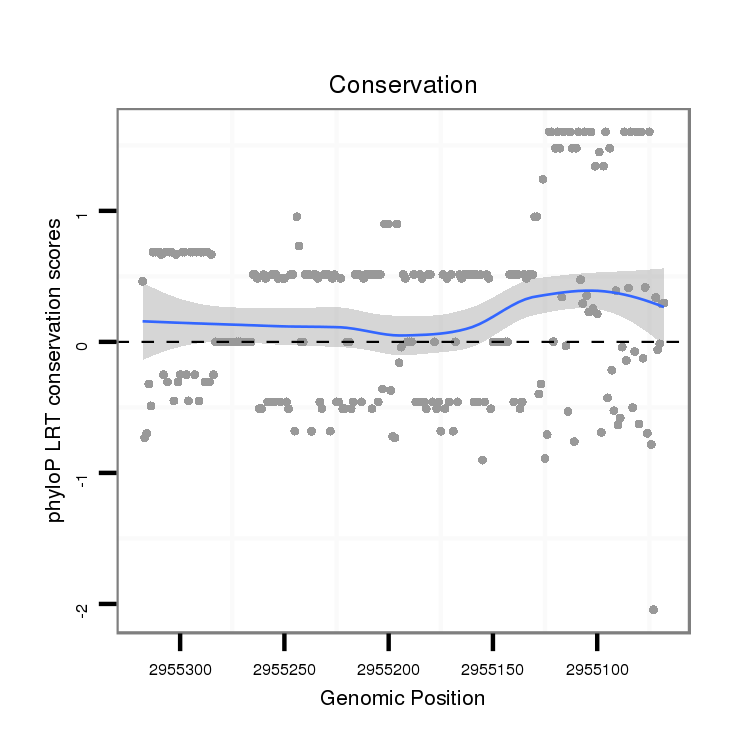
ID:dvi_13024 |
Coordinate:scaffold_12970:2955118-2955268 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ18987-cds]; exon [dvir_GLEANR_3802:2]; intron [Dvir\GJ18987-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGCTTTTTACTTATTTCTGTTGCAATTCAAATAGGTTTTTTTTTAATATTGCATAGTATGCGATTCCGGTTAAGATTTTATATAGCTTTTCGTACGATACACAAAGCTAATATCAATAATGGTTGACGTCTTAAGCTTAAGTTTATGCATTCACAATTTGTCACAAGTTGTTGAACTTATATCTGAATTTTTTAATTGCAGCTAGTGGCAACTATTGGGTACTTGGTGCACTGCTGATAATACTGACCGCC **************************************************........(((((((....(((((((......(((((((..(((...)))..)))))))...(((((....))))).)))))))...((((((((.......((((((((...))))))))))))))))..............))))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060684 140x9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
V116 male body |
SRR060663 160_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
M047 female body |
SRR060666 160_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
M061 embryo |
SRR060672 9x160_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060667 160_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
V053 head |
SRR060657 140_testes_total |
V047 embryo |
SRR060665 9_females_carcasses_total |
M027 male body |
SRR060659 Argentina_testes_total |
SRR060673 9_ovaries_total |
SRR060660 Argentina_ovaries_total |
SRR060683 160_testes_total |
SRR060689 160x9_testes_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................GGGAATCCGGTTAAGATT.............................................................................................................................................................................. | 18 | 2 | 5 | 36.20 | 181 | 34 | 27 | 2 | 13 | 21 | 10 | 3 | 10 | 10 | 9 | 6 | 6 | 5 | 1 | 5 | 3 | 3 | 1 | 3 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AAGGGAATCCGGTTAAGATT.............................................................................................................................................................................. | 20 | 3 | 9 | 20.11 | 181 | 15 | 30 | 54 | 29 | 10 | 11 | 3 | 2 | 0 | 1 | 5 | 4 | 3 | 0 | 0 | 3 | 0 | 1 | 1 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| ......................................................................................................................................................................................................................TTGGGTACTTGGTGCACTGCT................ | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GGGAATCCGGTTAAGATTC............................................................................................................................................................................. | 19 | 3 | 20 | 1.80 | 36 | 8 | 2 | 2 | 0 | 5 | 0 | 2 | 1 | 3 | 2 | 1 | 0 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................GTAAGGGAATCCGGTTAAGA................................................................................................................................................................................ | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................GAATGGGAATCCGGTTAAGATT.............................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................ATAGAAAGCGAATCCGGTTAAG................................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GCGAATCCGGTTAAGATT.............................................................................................................................................................................. | 18 | 1 | 2 | 1.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AAGCGAATCCGGTTAAGATTC............................................................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AAGGGAATCCGGTTAAGATTT............................................................................................................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TGAACTTATCTCTGCATT.............................................................. | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................CTTGGTGCAGTTCTGAT............. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................GGACTCCGGTTAAGATTC............................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................ATTATCAATAATGGTTCACA........................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACGAAAAATGAATAAAGACAACGTTAAGTTTATCCAAAAAAAAATTATAACGTATCATACGCTAAGGCCAATTCTAAAATATATCGAAAAGCATGCTATGTGTTTCGATTATAGTTATTACCAACTGCAGAATTCGAATTCAAATACGTAAGTGTTAAACAGTGTTCAACAACTTGAATATAGACTTAAAAAATTAACGTCGATCACCGTTGATAACCCATGAACCACGTGACGACTATTATGACTGGCGG
**************************************************........(((((((....(((((((......(((((((..(((...)))..)))))))...(((((....))))).)))))))...((((((((.......((((((((...))))))))))))))))..............))))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060660 Argentina_ovaries_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................CCGACGATCACCGTTGATAAA.................................. | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ........................................................................................TAGCATGCTATGTGTTAC................................................................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................ACCACATGACGAATGTTATG........ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:2955068-2955318 - | dvi_13024 | TGCTTTTTACTTATTTCTGTTGCAATTCAAATAGGTTTTTTTTTAATATTGCATAGTATGCGATTCCGGTTAAGATTTTATATAGCTTTTCGTACGATACACAAAGCTAATATCAATAATGGTTGACGTCTTAAGCTTAAGTTTATGCATTCACAATTTGTCACAAGTTGTTGAACTTATATCTGAAT----------TTTTTAATTGCAGCTAGTGGCAACTATTGGGTACTTGGTGCACT---GCTGATAATACTGACCGCC |
| droMoj3 | scaffold_4027:3061-3118 + | TTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATTGCAGCTTGTGGCAGCTATT------TTGGAGTCCTCTTGCTACTGATACTGTGTGCC | |
| droGri2 | scaffold_15081:868146-868340 + | TTATG------------------------------------------------TAGATTGTAACTCTGGCAAATA---TATCTAGTATTTAGTGTGTA--TTAAAACTAAAATTATTAAAACTCGAC---TCGAGTATAG-ACGAAGTAG-TACAATTTATTAGGAGA--------TTGTAATTGAATATTTGTTATATTACTACTTGCAGGTCCTGTCAACTATTGGCTGCTTGCAGCAAT---GCTGATAATATTCACAACC | |
| droWil2 | scf2_1100000004515:1429167-1429270 - | TACGCTTTACCTTTTGCAATTAAAACTAAATTTGA--------------------------------------------------------------------------------TTAATTCTCA-----------------------------------------------------------------------------TCTTTACAGCTCCTGCCAAGTGTTTGCTACTAGCCGGTTT---ATTTCTTAGCCTAACCAGC | |
| dp5 | XL_group3a:173616-173680 - | TTTCG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTGCAGCTCTTGTCAAGTGCTGGCTCCTGGGTTTCCTTCTACTGCTCACAGTTTCCGCG | |
| droPer2 | scaffold_39:561267-561331 + | TTTCG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTGCAGCTCTTGTCAAGTGCTGGCTCCTGGGTGTCCTTCTACTGCTCACAGTTGCCGCG | |
| droEle1 | scf7180000491006:450484-450564 - | AACTG---------------------------------------------------------------------AT------------------------------------------------------------------------------------------------------------------TTTCTATATTTTTTCTTACAGCTCCTGCCAAGTGCTGGCTCCTCGGTGTCCTGCTTCTACTCACAATTGCCGCC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 05/16/2015 at 07:45 PM