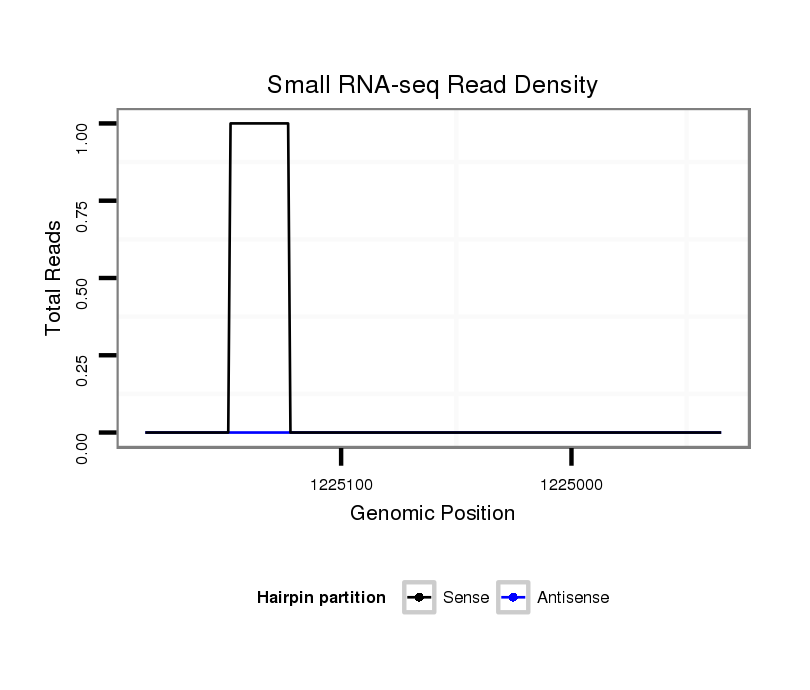
ID:dvi_12788 |
Coordinate:scaffold_12970:1224985-1225135 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_3869:2]; CDS [Dvir\GJ19055-cds]; intron [Dvir\GJ19055-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------###########--------------------------------------- GTAACAGCGCTTATTTATGTAAGCACACACACAGTGGATAGCTTTGCATGTGTGCGTGTGCGCTACCTCAACATTTCAATAAACTGCGATCTGCAATTTCTTTCATGTTTTTTTTTTTATTATATTTTCAATTTTTTTTGTTGTCTTCTGAGCTTGTCGCTCATTCAACTCCATAAATTTGTCATTCATTTTAATTTCCAGCTTATCGCTAAATTGTTGCGATAACAGCGTGCAGTTCTTACATTACGCAT **************************************************.((((....))))......(((((......(((.(((.....))).))).....)))))...................(((.(((....((((.....(((((.....)))))..)))).....))).)))....................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
SRR060670 9_testes_total |
GSM1528803 follicle cells |
M047 female body |
SRR060676 9xArg_ovaries_total |
SRR060668 160x9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................ATAGCTTTGCATGTGTGCGTGTGCGC............................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TAAACTGAGATCTGCCGTTTCTT..................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................GCCTTGCATGTGTGCGTGT................................................................................................................................................................................................ | 19 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................GTCATTCATTTTAGATCCCA................................................... | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................CTGCAGTTCTTACAAAACGC.. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................TTATTTATTATATTTTCAAT....................................................................................................................... | 20 | 1 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................TTCAACTCCATAAAATTGTTT................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CATTGTCGCGAATAAATACATTCGTGTGTGTGTCACCTATCGAAACGTACACACGCACACGCGATGGAGTTGTAAAGTTATTTGACGCTAGACGTTAAAGAAAGTACAAAAAAAAAAATAATATAAAAGTTAAAAAAAACAACAGAAGACTCGAACAGCGAGTAAGTTGAGGTATTTAAACAGTAAGTAAAATTAAAGGTCGAATAGCGATTTAACAACGCTATTGTCGCACGTCAAGAATGTAATGCGTA
**************************************************.((((....))))......(((((......(((.(((.....))).))).....)))))...................(((.(((....((((.....(((((.....)))))..)))).....))).)))....................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060678 9x140_testes_total |
V053 head |
SRR060687 9_0-2h_embryos_total |
SRR1106729 mixed whole adult body |
SRR060682 9x140_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060670 9_testes_total |
SRR060666 160_males_carcasses_total |
SRR060667 160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................CGTGAAGACTGTAATGCGTA | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................GTCACCTAGCGTAACGTACCCA...................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................GCACCGGCGATAGAGTTGTA................................................................................................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................TCGTGGGTGTGGTACCTATC.................................................................................................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GTGTTTGTCACCTCTCGATAC............................................................................................................................................................................................................. | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................TTACAGAAAGTACCAAAAA.......................................................................................................................................... | 19 | 2 | 18 | 0.11 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................TAGGTCGAATAGCAATTT...................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TGTAGGTCGAATAGCAATTT...................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................AAGTTCAAGAAAGTACCAAA............................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................GTTATTGTCGATCGTCAAGA............ | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................AAGTTATTTGACTGTAGA............................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................TTAAAGGTCGAATACAGA......................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:1224935-1225185 - | dvi_12788 | GTAACAGCGCTT---A--T---------------------------TTATGTAAGCACACACACA-GTGGATAGCTTTGCATGTGTGCGTG-----------------TGCGCTACCTCAACA-TTTC--------------------AATAAACTGC------GAT-----CTGCAATTTCTTTCATGTTTTTTTTTTTATTATATTTTCAATTTTTTTTGTTGTC-TTCTGAGCTTGTCGCTCATTCAACTC----------------------------------CATAAATTTGTCATTCATTTT--AATTTCCAGCTTATCGCTAAATT-----------------GTTGCGATAACAGCGTGCAGTTCTTACATTACGCAT |
| droMoj3 | scaffold_6308:1169792-1170037 + | GTTTG-------------CTG-----------------------------TTATGCGCACACTCA----------------------TGCACTG--------------TTAGCTACCTCAACA-TTTC-TACTCTGCCCATCTGCTGCTGCTGTTATC------GCT-----TTAATCTTTTTGTT---------------------GTTCTTTTTTTTTTGT-GAC-TTTTGAACGTGTTGCCCATTCAACCCGCTGTTGTTCATGGCTGAAATTATTATTTATTTGCATATATTTGTCATACGCTCTGCCATTGCAAACTTATCGCCAAATT-----------------GTTGCGATAAG----------TCTTATATTACGCAT | |
| droGri2 | scaffold_15081:1811134-1811368 - | GTACGAATGT-----ATGT-----------------------------ATGTATGTCCATATGTATGTATGTATGTATGCATGTGTGTGCATTGTACGTTTATTCTTTTTCACTACCTCAACA-TTTGTGGCTCTTTCTGCCTGCTTTATCTGCGCTC------GTCTCTTGTCAGTCTT-------------------------------------------------------CTTGTCGCCCATTCAACTT-----------------------------------------TTGTCATTCA-------------GGTTTGTCACTCAATTTTAATGAAAACTTTAAAGCTGCGATAAGATCAGACATTTCTT--ATTACGCAT | |
| dp5 | XL_group1a:2179065-2179225 + | CCAAGATCGGTT---G--TCGTTCAGCGCAATTTTTACAACAGATTTGCTTTTT--TTGCTTGCTTGT------------GTGTGTGTGTGTTTCGG--------TTTTT--------CGCTG-CT--------------------GTAGTAATTCTC------GGT-----TTTGGCGTTTTTTGGTTTTTTTTTGTTTTTTGTATTTTTTGTTTTTTTCGTTGTT-T-------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453912:1460862-1461005 + | CTGCTGATGCTGCAGATGT---------------------------TGCTGTTG--CCACATGCCTTGAGCTGT---------------TGCGTCAG--------TTGCT--------CGCTAACG--------------------ATGACAATTCCATAAACATGT-----TTTCGGTTTCCTTCCTGTTTCGTTTGTTCTTCAACTTTCCTCTTTCTTTTTGGTTTT-------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
Generated: 05/16/2015 at 09:22 PM