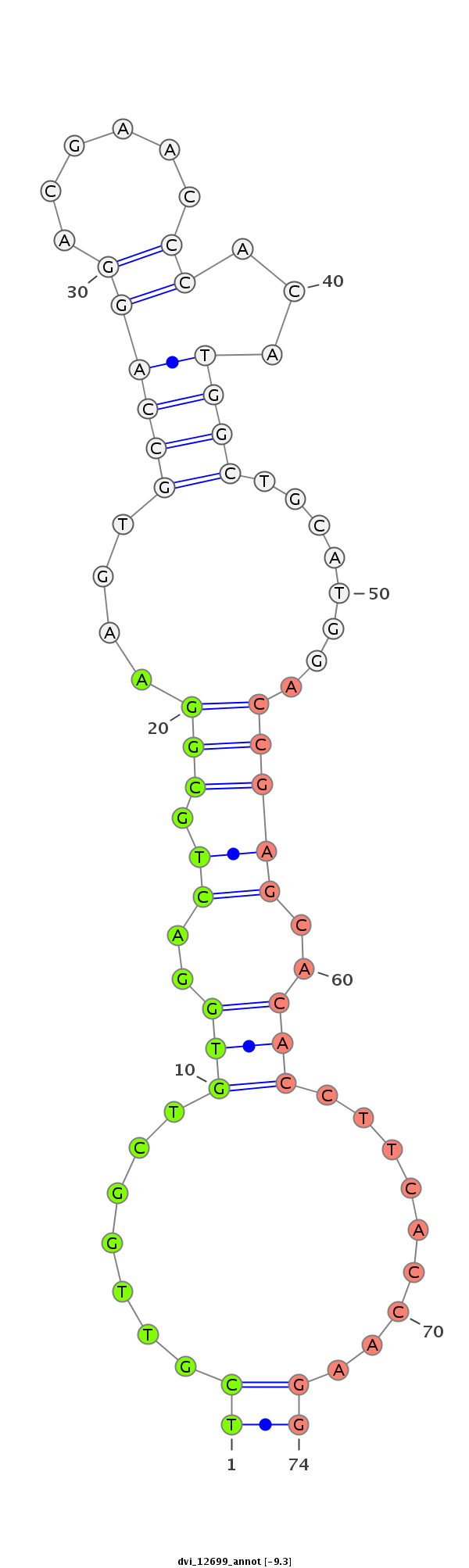

ID:dvi_12699 |
Coordinate:scaffold_12967:719661-719811 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.7 | -16.6 | -16.5 | -16.4 |
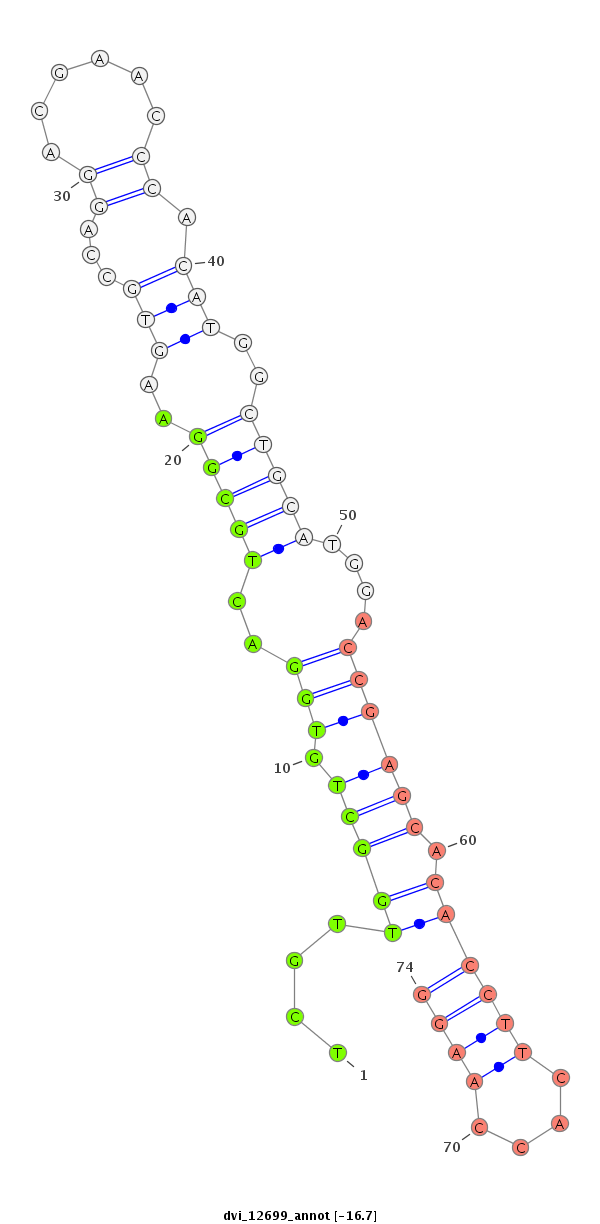 |
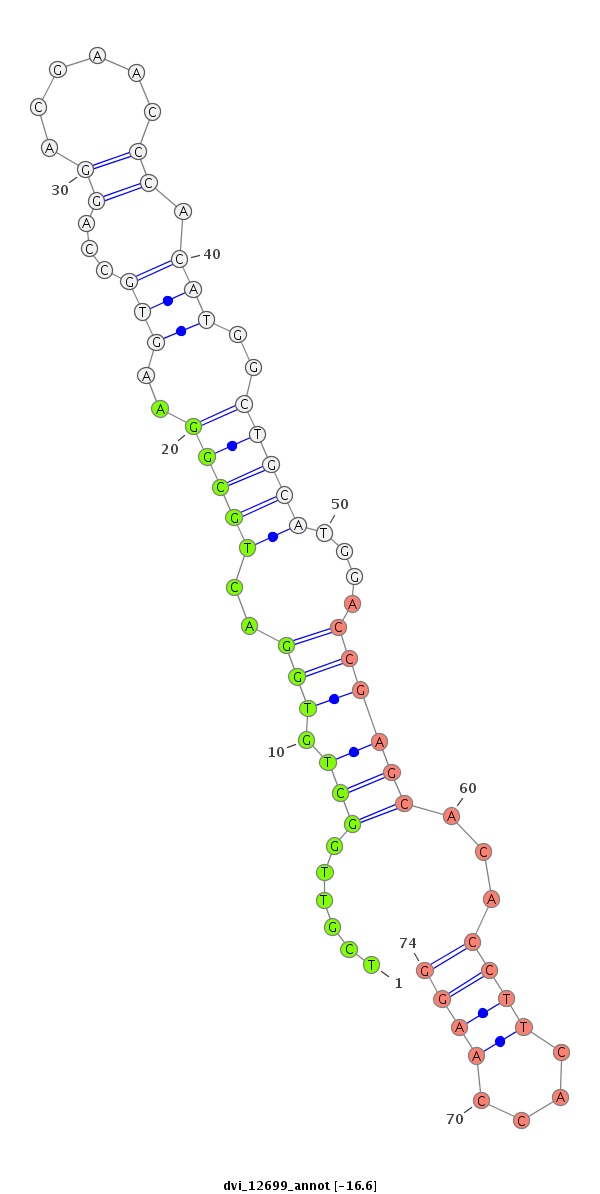 |
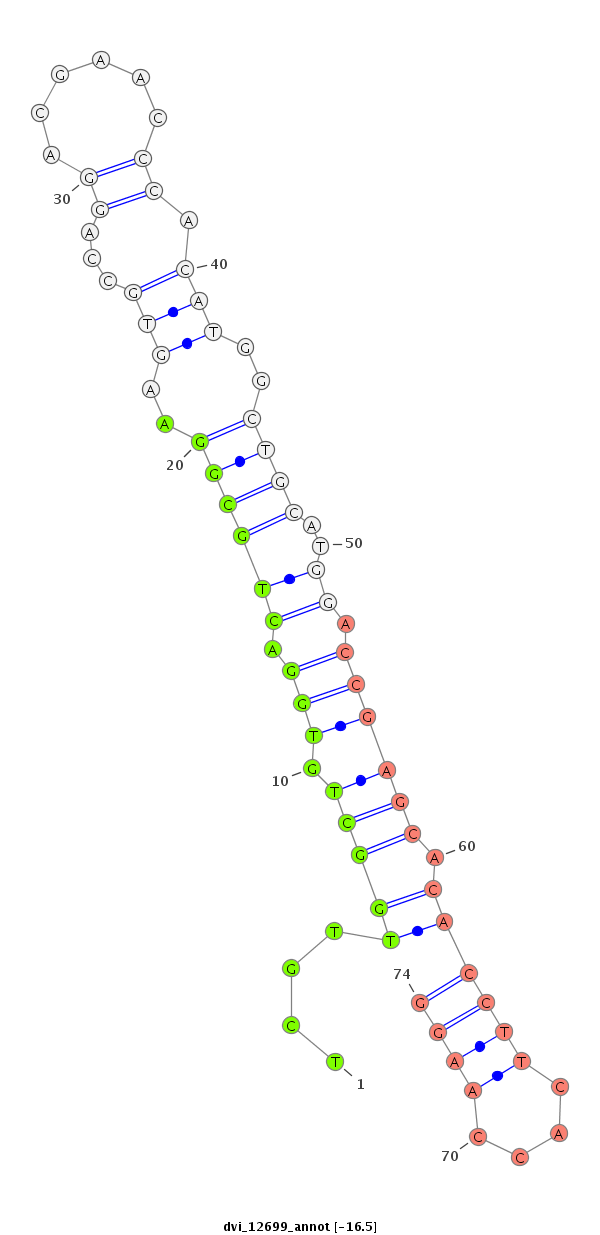 |
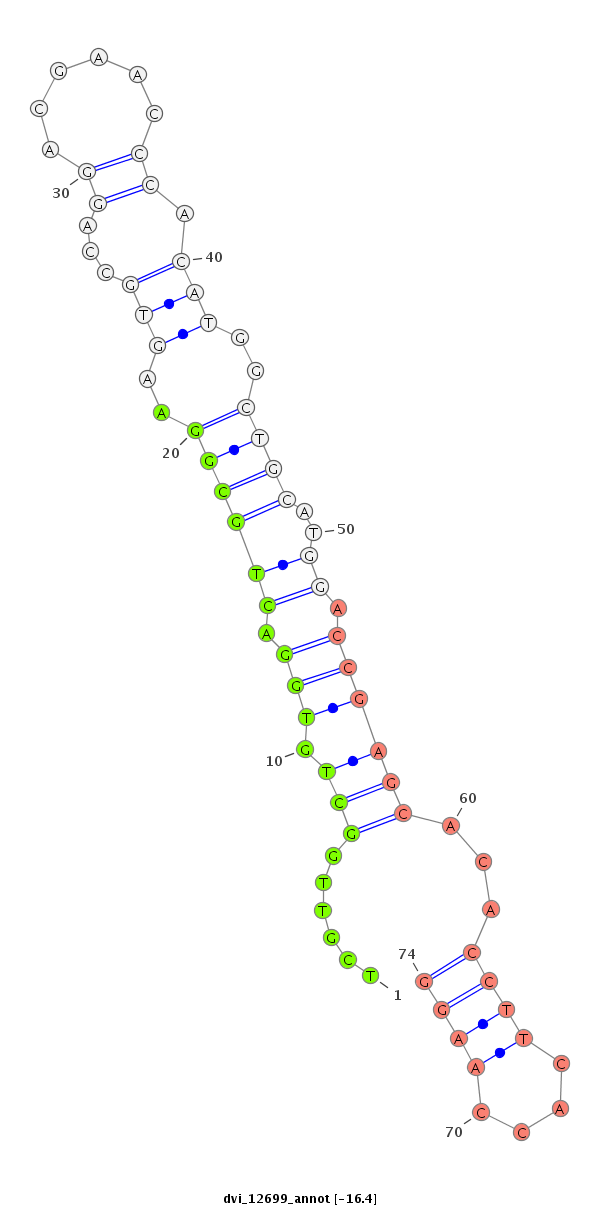 |
exon [dvir_GLEANR_15233:1]; CDS [Dvir\GJ14932-cds]; intron [Dvir\GJ14932-in]
No Repeatable elements found
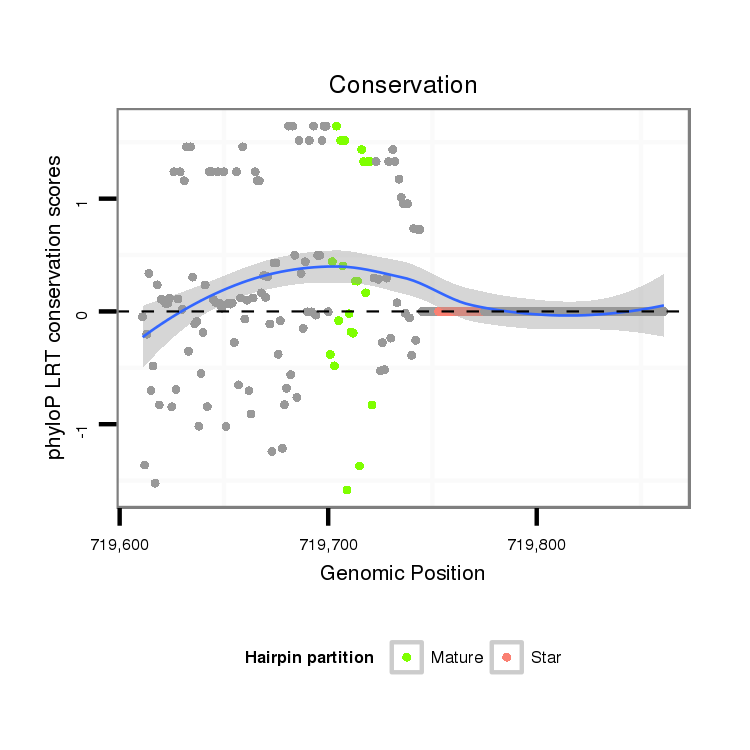
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CTGCTACCATTCATCTCGACGAATCTGCACGAATGCTGGACGAACTAATGGTAAACGCACTTCCTACTGTTTATTGCCGACAATTTCAACTCGTTGGCTGTGGACTGCGGAAGTGCCAGGACGAACCCACATGGCTGCATGGACCGAGCACACCTTCACCAAGGCAAGCCCTTCAATACCAAGTCGGACTACATGGGAGGTGTCCCAAATGTACACACAAAGTGACACCCAGCATATTGTCAGCAGTTCTA ******************************************************************************************((.......(((..((.(((....((((((......))...))))........)))))..))).........))*************************************************************************************** |
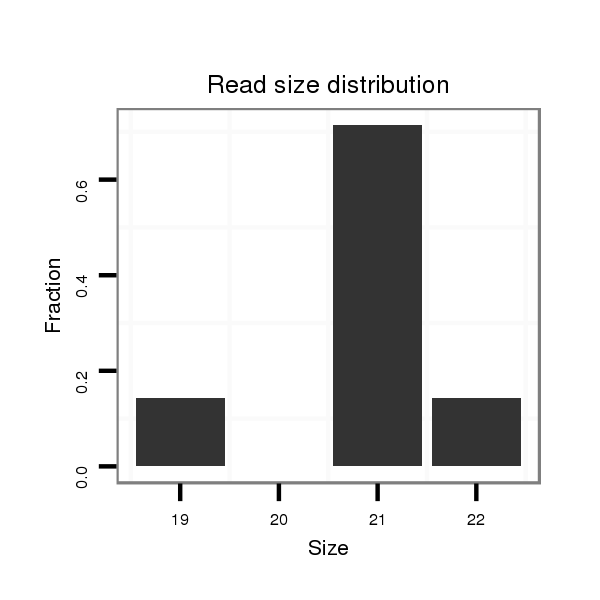
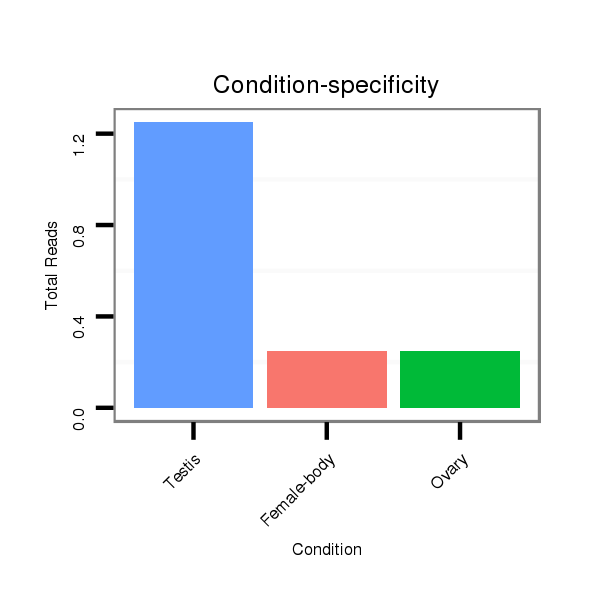
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060670 9_testes_total |
M047 female body |
SRR060675 140x9_ovaries_total |
SRR060680 9xArg_testes_total |
SRR060687 9_0-2h_embryos_total |
V053 head |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................TCGTTGGCTGTGGACTGCGGA............................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................ACCGAGCACACCTTCACCAAGG....................................................................................... | 22 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................CCGAGCACACCTTCACCAAGG....................................................................................... | 21 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................CCGAGCACACCTTCACCAA......................................................................................... | 19 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCTCGGTGGCTGTGGACT................................................................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................TGGACGGACTATTGGTAAG.................................................................................................................................................................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................CTGCGGAAGTGCATGGCCG................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TGGCAGCATCGACCGACCA..................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................CTGCCAGAATGCTGCACGA................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GACGATGGTAAGTAGAGCTGCTTAGACGTGCTTACGACCTGCTTGATTACCATTTGCGTGAAGGATGACAAATAACGGCTGTTAAAGTTGAGCAACCGACACCTGACGCCTTCACGGTCCTGCTTGGGTGTACCGACGTACCTGGCTCGTGTGGAAGTGGTTCCGTTCGGGAAGTTATGGTTCAGCCTGATGTACCCTCCACAGGGTTTACATGTGTGTTTCACTGTGGGTCGTATAACAGTCGTCAAGAT
***************************************************************************************((.......(((..((.(((....((((((......))...))))........)))))..))).........))****************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
GSM1528803 follicle cells |
SRR060682 9x140_0-2h_embryos_total |
SRR060675 140x9_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060656 9x160_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060660 Argentina_ovaries_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060670 9_testes_total |
SRR060673 9_ovaries_total |
SRR060674 9x140_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060687 9_0-2h_embryos_total |
V116 male body |
SRR060671 9x160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................CTGGCTCGTGTGGAAGTGGTTCCGT..................................................................................... | 25 | 0 | 4 | 1.50 | 6 | 1 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................GTGGGTCGTATAACAGTCGTCAAGAT | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TTCCGTTAGGGAAGTTATG........................................................................ | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CCTGGCTCGTGTGGAAGTGGTTCCGTA.................................................................................... | 27 | 1 | 4 | 0.75 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................GCTCGTGTGGAAGTGGTTCCGT..................................................................................... | 22 | 0 | 4 | 0.75 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CCTGGCTCGTGTGGAAGTGGTT......................................................................................... | 22 | 0 | 4 | 0.50 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TGGCTCGTGTGGAAGTGGTTC........................................................................................ | 21 | 0 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................CTGGCTCGTGTGGAAGTGGTT......................................................................................... | 21 | 0 | 4 | 0.50 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TGGCTCGTGTGGAAGTGGTTCCGT..................................................................................... | 24 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................GGGGTCGTGTGGAAGTGGTTAC....................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................CTGGCCTGGCTCGTGTGGAAGTG............................................................................................ | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GGCTCGTGTGGAAGTGGTTCCGT..................................................................................... | 23 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................TCCTGGCTCGTGTGGAAGTG............................................................................................ | 20 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GGGTCGTGTGGAAGTGGTTA........................................................................................ | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................CTGGCTCGTGTGGAAGTGGCTCCGT..................................................................................... | 25 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................GGTCGTGTGGAAGTGGTT......................................................................................... | 18 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................CTGGCTCGTGTGGAAGTGGT.......................................................................................... | 20 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TCTCCTGGCTCGTGTGGAAGTGGTTCCGT..................................................................................... | 29 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CCTGGCTCGTGTGGAAGTGGTTCC....................................................................................... | 24 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CCGGGGTCGTGTGGAAGTGGTTA........................................................................................ | 23 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CTTCACAGTCCTGCTTGGGT.......................................................................................................................... | 20 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CTGCTTAGAACCTGCTTGA............................................................................................................................................................................................................. | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................................TGGATTTACTTGTGTGTTTCA............................ | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................CCGGGGTCGTGTGGAAGTG............................................................................................ | 19 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................TCAGTTAGGGAAGTTATG........................................................................ | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................TGCTTACGACATGGTTAATT........................................................................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................CGGGGTCGTGTGGAAGTG............................................................................................ | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12967:719611-719861 + | dvi_12699 | CTGCTACCATTCATCTCGACGAATCTGCACGAATGCTGGACGAACTAATGGTAAACG--------CACTTCCTACTGTTTATTGCCGACAATTTCAACTCGTTGGCTGTGGACTGCGGAAGTGCCAGGACGAACCCACATGGCTGCATGGACCGAGCACACCTTCACCAAGGCAAGCCCTTCAATACCAAGTCGGACTACATGGGAGGTGTCCCAAATGTACACACAAAGTGACACCCAGCATATTGTCAGCAGTTCTA |
| droWil2 | scf2_1100000004947:266624-266753 - | CACCGAGCAAACACATCGACGAATTCTGCAGCATACTGGATGAATTAGTGGAGGACGCCAGAACGCACACGCCAACCATCATAGCCGGCGACTTCAATACATGGGCAACGGAATGGGGTAGCCAAAGGAC--------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_213:27522-27591 - | ACAC-------------------------------------------------------------------CCTACGCTCATAGCCGGCGATTTCAACGCTTGGGCGACGGAGTGGGGATGCCCGAGAACGAACCCA-------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13469:171664-171734 + | TGCGAGACG------------ACTTTAGAGG-----------------T--------GGTGAAAGCACTTCCCAAGTTTTACGGGGAACAAGCCCAATATGTTGGCTG------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453163:4583-4724 - | CTCCATCGCTAAGCCTGGAGGAGTTCACAGGTATACTTGACTTAATGGTCAGAGACGCGAGAGACAAGTCACCATGCTTAATCGCCGGCGATTTCAACGCATGGGCGATCGACTGGGGTAGCGTCAGTACAAACTCGAAAGG--------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000413131:10188-10319 - | CCCCGAGCGTACGCATTAACGAGTTCAGGGCAATAATGCAGGAGATTGTCGACGACGCACGCGGCAGGTCTTCCATCCTCATCGCAGGCGATTTCAACGCATGGTCCACAACATGGGGCAGCGCCTGTACGA------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/16/2015 at 08:32 PM