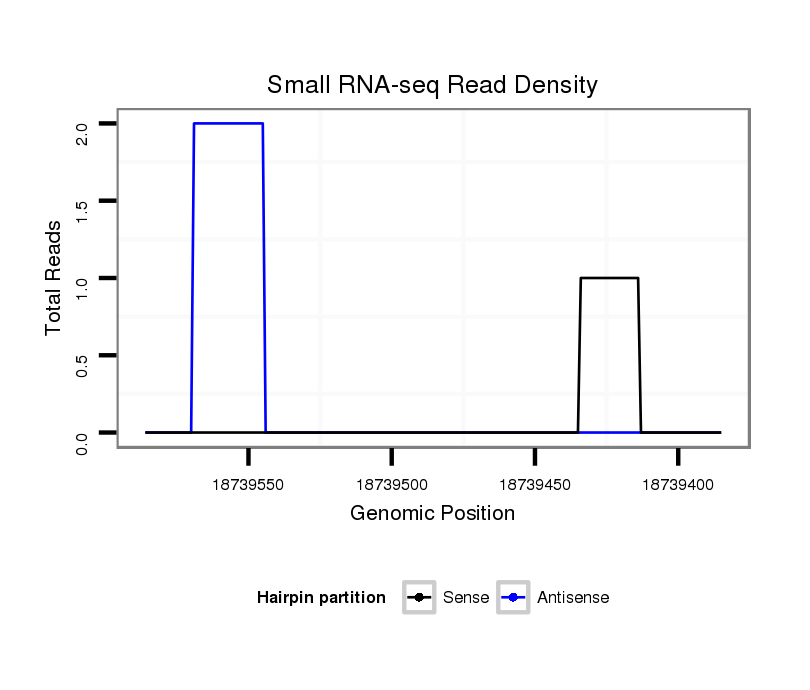
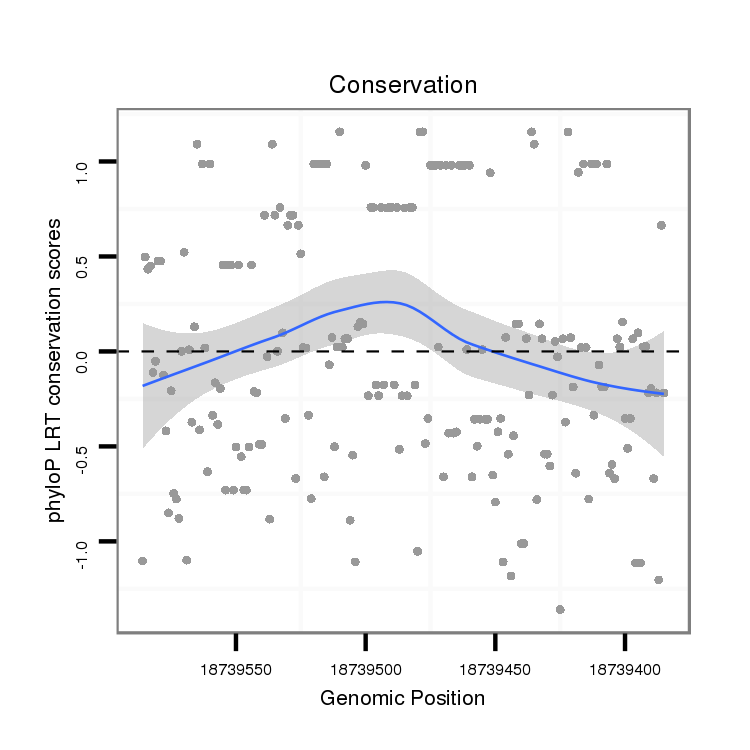
ID:dvi_12495 |
Coordinate:scaffold_12963:18739435-18739536 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_503:4]; exon [dvir_GLEANR_503:3]; CDS [Dvir\GJ19777-cds]; CDS [Dvir\GJ19777-cds]; intron [Dvir\GJ19777-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------------------------################################################## AAGAGGAAGGACTACAAGCTCGTTCCAAGACGCGCCGCTCCACAAGATCGGTAAGGGTTGCAACTGTTTATTCATTTAGCATACTATTTTATTAGTATACTCTTTGCATACTATTTTATAAGTAATTTATATTTGTCCCATTAATTAATTAGCTGGGCTCCCGAAAGAGTAAATTATCGACCGGAAGAGGTGTTTCGCCCGG **************************************************....(((..((((.(((..((.((...((.((((((......)))))).))...))..((((........))))))..))).))))))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
M028 head |
SRR060678 9x140_testes_total |
SRR060662 9x160_0-2h_embryos_total |
V116 male body |
SRR060663 160_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
V053 head |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................GGGTAGCGACTGTTTATT.................................................................................................................................. | 18 | 2 | 4 | 2.50 | 10 | 5 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................ATCGGTTAGGGTGGCAACT......................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................CTGGGCTCCCGAAAGAGTAAA............................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...ACGAAGGACTTCAAGCGCGTT.................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................GGGGTAGCGACTGTTTATT.................................................................................................................................. | 19 | 3 | 14 | 0.43 | 6 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| ........................................................................................................................................................CTGGGCTCAGGAAATAGTAA.............................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................TATCGACCACAAGCGGTGTT........ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................GGGTAGCGACTGTTTATTA................................................................................................................................. | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................GGGGTAGCGACTGTTTAT................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TTCTCCTTCCTGATGTTCGAGCAAGGTTCTGCGCGGCGAGGTGTTCTAGCCATTCCCAACGTTGACAAATAAGTAAATCGTATGATAAAATAATCATATGAGAAACGTATGATAAAATATTCATTAAATATAAACAGGGTAATTAATTAATCGACCCGAGGGCTTTCTCATTTAATAGCTGGCCTTCTCCACAAAGCGGGCC
**************************************************....(((..((((.(((..((.((...((.((((((......)))))).))...))..((((........))))))..))).))))))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060672 9x160_females_carcasses_total |
M047 female body |
SRR060673 9_ovaries_total |
SRR060666 160_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060671 9x160_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060665 9_females_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................CGAGCAAGGTTCTGCGCGGCGAGGT................................................................................................................................................................ | 25 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TAATAGTTGTACTTCTCCAC.......... | 20 | 3 | 14 | 1.43 | 20 | 1 | 0 | 0 | 9 | 1 | 3 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 0 |
| ............................................................................................................................................................................TAATAGCTGTACTTCTCCAC.......... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TCTACTCCACAAAGCGGGA. | 19 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................CCGACGTTGACAACTAAGT................................................................................................................................ | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................TGGTGTGCTAGCCATTCACA................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................CGGCGATGTGATATAGCC....................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:18739385-18739586 - | dvi_12495 | AAGAGGAAGGACTACAAGCTCGTTCCAAGACGCGCCGCTC-CACAAGATCGGTAAGGGTTGCAACTGTTTATTCATTTAGCATAC----T-ATTTTATTAGTATA--CTCTTTGCATA-----CTATTTTAT---AAGTAATTTATATTTGTCCCATTAATTAATTAGCTGGGCTCCCGAAAGAGTAAATTATCGACCGGAAGAGGTGTTTCGCCCGG |
| droMoj3 | scaffold_6500:24422158-24422305 + | TAGAGCAAGCACCCATTTTACG-------TCGAGCAACAAGAGCAAGCTCTGT--AAGTTTC------------TTTTAGCGCCC----T-AT---------------------AATT-----TTATTTCAA---ATATAATTCGAGTCCGGATTCTCCATTTCCTAGATGGGCTCCAGAAG---------------CAGAAAAAGCACATTGATTGA | |
| droGri2 | scaffold_15252:6656527-6656672 - | CAGAGGAACGTGAAAAATTACGATCAAGAAT-------------GTCCTCAGT--GAGTTTCAACCATTTAATCAGCTGATTTGCGAACTTA----------------------TATT-----T------------------------------TGTCGTTTCTTAAGATAAAGACCGATAGGCGTAAGTCATCGATGAAGAAGAAGGGAACAACGGG | |
| droWil2 | scf2_1100000004516:1631621-1631641 - | AGGAAGAAGAGCAGCGAATTC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396430:499423-499440 - | GAGGCGAAGATTCACAAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4845:5604347-5604523 + | AAAACAGGTACTCTAAAT-TGGTTTAAAGT-------------------GGG--AGG------GTTCTTTACTAAATTAGCAGAT----C-TTATTGATGGTACAAAGACATTAAATATCAAACTATCTAATGGAAAATAAATAAAAATTGTTTCAGA-CAAAATAAGCAGCCGAAGTGGAGCAGCAGCTTAGGCAATCGAGGCGGTGTAT------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:29 AM