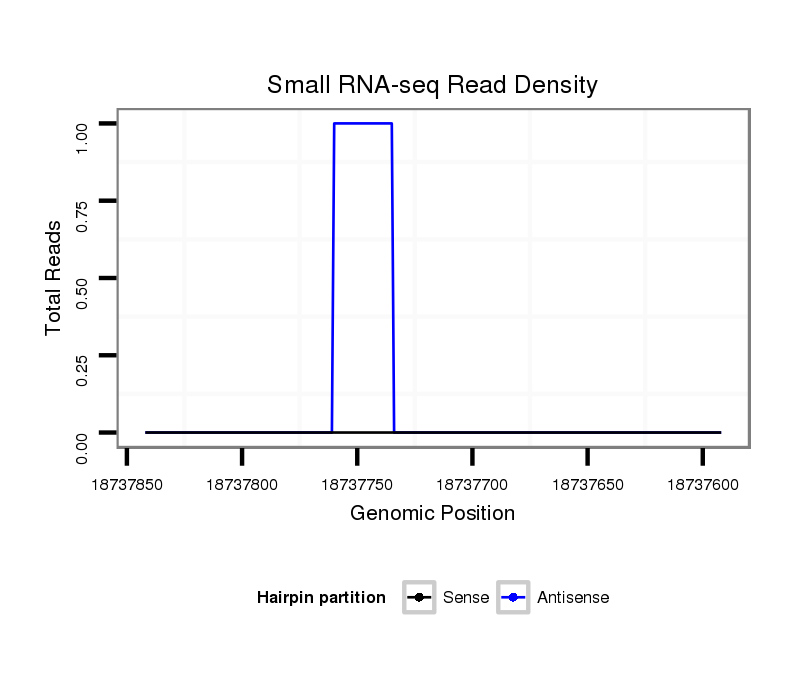
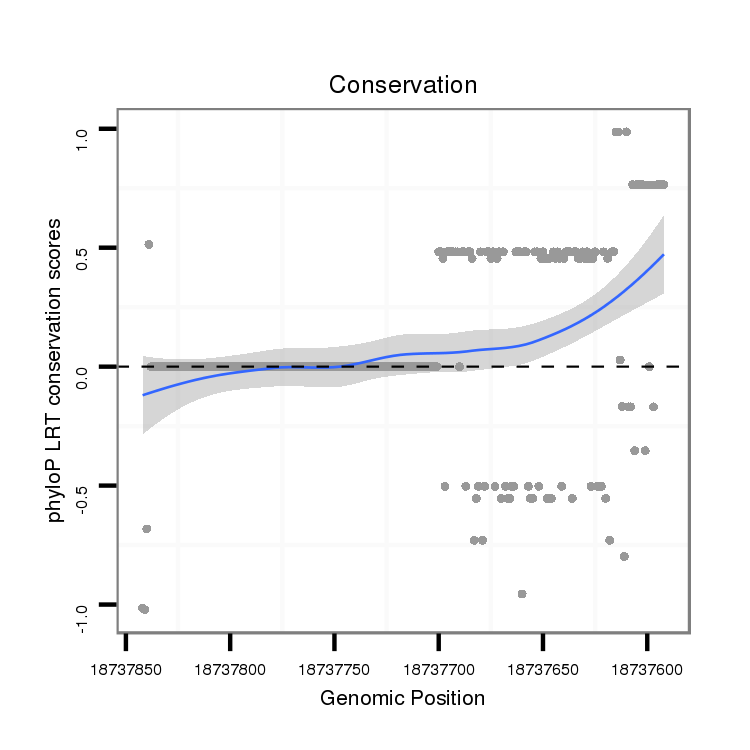
ID:dvi_12493 |
Coordinate:scaffold_12963:18737642-18737792 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_504:2]; CDS [Dvir\GJ19784-cds]; intron [Dvir\GJ19784-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------##############################-------------------- GTATTATTACAATTTCAAAAGATTTACATCTATTAGAAATTTTAAAGATTCTGAAAATATTAGGTTCACAAAGTACAATAAACGTCAGCTCCTTAAGTCACAGTTTTAAGGTCACTGAGAGTTACAGTTTTAAAGTCCCAAAATCAAATAATCAAGTAGCTTATTTTCTACTAATTACATATGTTTTAATGCTGTGAACAGAGTATAAAGCAGTCGGTTGCAACCAATTAGGAGCTTAATACCAGGCTTTT **************************************************((((.....))))(((((((..(((....(((((((((..(((((((.......)))))))...))))(((((((..............................)))))))...................)))))...))))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060670 9_testes_total |
V047 embryo |
SRR060655 9x160_testes_total |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................TAGGTGCTTAATACCGCGCTTT. | 22 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTAATACCCTGCTTT. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTCATACCGGGCTAT. | 22 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTAATACAGCGCTTT. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAACTTAATACCAGGCTAT. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTGATACCGTGCTTT. | 22 | 3 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAACTTAATACCCGGCTAT. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTAATACCGTGCTTT. | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTCATACCGCGCTTT. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAACTTACTACCGGGCTTT. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTCATACCGGGCT... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTCATACCTGGCTCT. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTAATAACGTGCTTT. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAACTTAATACCTGGCTATT | 23 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTCATACCCGGCTCT. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTCATACCGCGCTT.. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAACTTAATACCGGGCTT.. | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTAATACCATGCTCT. | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TATGAACTTAATACCGGGCTTT. | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGGAGCTTAATACCGTGCTTC. | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................TTACATATGTTTGAAGGATGTG....................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................TAGGAGCTTGATACCGGGCTCT. | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGTAGCTTAATACCAGCCA... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................ATATGTCTTTATGCTGGGAA..................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
CATAATAATGTTAAAGTTTTCTAAATGTAGATAATCTTTAAAATTTCTAAGACTTTTATAATCCAAGTGTTTCATGTTATTTGCAGTCGAGGAATTCAGTGTCAAAATTCCAGTGACTCTCAATGTCAAAATTTCAGGGTTTTAGTTTATTAGTTCATCGAATAAAAGATGATTAATGTATACAAAATTACGACACTTGTCTCATATTTCGTCAGCCAACGTTGGTTAATCCTCGAATTATGGTCCGAAAA
**************************************************((((.....))))(((((((..(((....(((((((((..(((((((.......)))))))...))))(((((((..............................)))))))...................)))))...))))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
M028 head |
M047 female body |
SRR060679 140x9_testes_total |
V047 embryo |
SRR060684 140x9_0-2h_embryos_total |
SRR060678 9x140_testes_total |
SRR060669 160x9_females_carcasses_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................TGGCTCTCAATGTCAGAA........................................................................................................................ | 18 | 2 | 3 | 17.33 | 52 | 49 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CCGATGACTCTCAATGTCAGAA........................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CCAATGGCTCTCAATGTCAGAA........................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CCGATGGCTCTCAATGTC............................................................................................................................ | 18 | 3 | 20 | 1.00 | 20 | 18 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................GCAGTCGAGGAATTCAGTGTCAAAAT............................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................GGGCTCTCAATGTCAGAA........................................................................................................................ | 18 | 3 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CGGCTCTCAATGTCAGAA........................................................................................................................ | 18 | 3 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AAGTGTTCCATGTTATTT......................................................................................................................................................................... | 18 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TGGTTAATCCTGCAACTATG......... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................TGGCTCTCAATGTCAGAACT...................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TCACGTTGGTTAATCCTC................. | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................CCGATGGCTCTCAATGTCA........................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................AAAGTGTTCCATGTTATT.......................................................................................................................................................................... | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................CAAAATTTCTGGGTT.............................................................................................................. | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................AGTTGGTCGAATAAAAGA.................................................................................. | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................TAGTTGGTCGAATAAAAG................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................TGGCTCTCAATGTCAGAAC....................................................................................................................... | 19 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TAGGGTCTTAGGTTATTAG.................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................TATTAGTTTATCGATTAAA..................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:18737592-18737842 - | dvi_12493 | GTATTATTACAATTTCAAAAGATTTACATCTATTAGAAATTTTAAAGATTCTGAAAATATTAGGTTCACAAAGTACAATAAACGTCAGCTCCTTAAGTCACAGTTTTAAGGTCACTGAGAGTTACAGTTTTAAAGTCCCAAAATCAAATAATCAAGTAGCTTATTTTCTACTAATTACAT-ATGTTTTAATGCTGTGAACAGAGTATAAAGCAGTCGGTTGCAACC--AATTAGGAGCTTAATACCAGGCTTTT |
| droMoj3 | scaffold_6500:24423630-24423724 + | AA--------------------------------------------------------------------------------------------------------------------------------------------ATCGAATAAT-AAATAGAACAGCTTCTGCTTACATTGTGATCTTCATATACTGAGTACAGGGTATTAAGCAGTCAGTCATATCAATAATTAAAA------------------ | |
| droGri2 | scaffold_15252:6655406-6655409 - | TCCT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004516:1630440-1630440 - | T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409447:617733-617757 - | AA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGGCAATTGAATAAC-GACTTTT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/17/2015 at 04:28 AM