
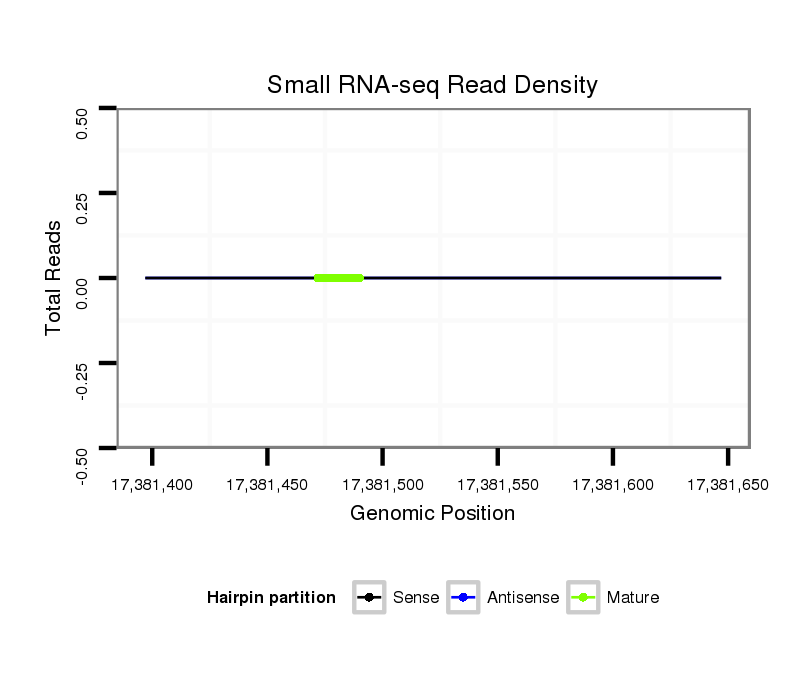

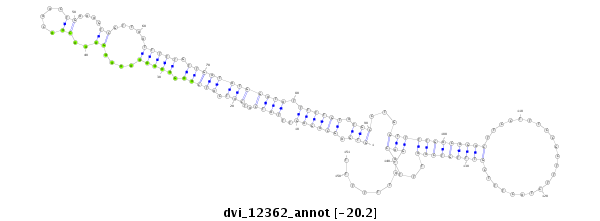
ID:dvi_12362 |
Coordinate:scaffold_12963:17381447-17381597 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.5 | -20.2 | -20.2 |
 |
 |
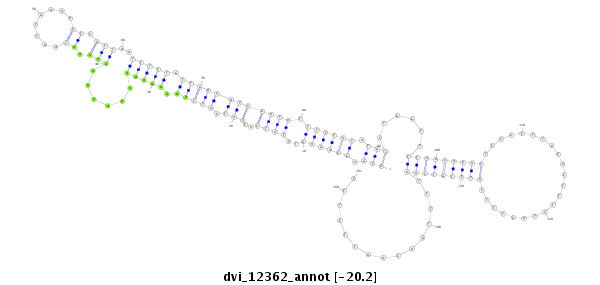 |
intron [Dvir\GJ17715-in]; exon [dvir_GLEANR_2286:8]; CDS [Dvir\GJ17715-cds]; intron [Dvir\GJ17715-in]
No Repeatable elements found
| mature | star |
| --------------####################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTTTTGTACACCAGGTCTGATAAAAAAAAAGGCGGTTGCCAAAGTCACAGGTGAGCAGAGTTTATCATCATCATCAAGAAAAAAAAAGAAACAACAACTGAAAGTCGTTAATTTTTGTTGATATGGATATTTTTGTATGCCTGGTTTTGACAGACTTCCTTTCCGCTTTCTACTTTCGTTTGTCAATTTTGACAATTTTTTGCTTCACGTTTTTATTTTATTTGGTTGTTATGCAAAAAAAAAAAAGGGAG **************************************************(((.((((((..((((..(((.(((((.(((((......(((.((........)).)))..))))).)))))))))))).)))))).)))...((((((((((((......................)))))))))....)))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
SRR060683 160_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060654 160x9_ovaries_total |
M047 female body |
SRR060655 9x160_testes_total |
SRR060679 140x9_testes_total |
SRR060689 160x9_testes_total |
SRR060677 Argx9_ovaries_total |
V116 male body |
SRR060680 9xArg_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................TTCTTATTTTATTGGGTTCTTATGCA................ | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TTCTTATTGTATTTGGTTTTTATGCA................ | 26 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GTCTTATTTTATTTGGTTTTTATGCAA............... | 27 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TATTTAGAAGTTATGCAAAAAAAAAAA...... | 27 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TTCTTATTTTATTTTGTTATTATGCA................ | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GTCTTATTTTATTTGGTTGTTCTGCA................ | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TTCTTATTGTATTTGGTTTTTATGC................. | 25 | 3 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................TGTTGATCTGGATCTTTTTGTC.................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TTCTTATTTTATTTGGTTTTTATGC................. | 25 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TTCTTATTTTATTTGGTTTTTAT................... | 23 | 2 | 10 | 0.30 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................AAGAAAAAAAAAGAAACAA............................................................................................................................................................. | 19 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AAGCAGAGTTTATCAACATCG.................................................................................................................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TTGTTATGCGAAAAAAAAAA...... | 20 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TTGTTATGCGAAAAAAAA........ | 18 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................AGCAGAGTTTATCAACATCGA................................................................................................................................................................................. | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TTTGGTTGTGATGCAAGAAAT.......... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................TTCTTATTTTATTTGTTTTTTATGC................. | 25 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................ATTTTATTTGGTTTTGATGC................. | 20 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TGATCATCAGGAAAAAAA...................................................................................................................................................................... | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................CTTTCTGCTTTCGTTGGT.................................................................... | 18 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CAAAACATGTGGTCCAGACTATTTTTTTTTCCGCCAACGGTTTCAGTGTCCACTCGTCTCAAATAGTAGTAGTAGTTCTTTTTTTTTCTTTGTTGTTGACTTTCAGCAATTAAAAACAACTATACCTATAAAAACATACGGACCAAAACTGTCTGAAGGAAAGGCGAAAGATGAAAGCAAACAGTTAAAACTGTTAAAAAACGAAGTGCAAAAATAAAATAAACCAACAATACGTTTTTTTTTTTTCCCTC
**************************************************(((.((((((..((((..(((.(((((.(((((......(((.((........)).)))..))))).)))))))))))).)))))).)))...((((((((((((......................)))))))))....)))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060663 160_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
V116 male body |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060672 9x160_females_carcasses_total |
M027 male body |
M028 head |
SRR060669 160x9_females_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
V047 embryo |
SRR060681 Argx9_testes_total |
M047 female body |
V053 head |
SRR060657 140_testes_total |
SRR060664 9_males_carcasses_total |
SRR060675 140x9_ovaries_total |
SRR060660 Argentina_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060678 9x140_testes_total |
M061 embryo |
SRR060676 9xArg_ovaries_total |
SRR060654 160x9_ovaries_total |
SRR060673 9_ovaries_total |
SRR060655 9x160_testes_total |
SRR060656 9x160_ovaries_total |
SRR060658 140_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......TGTGGTACAGACTAGTTT.................................................................................................................................................................................................................................. | 18 | 2 | 4 | 292.75 | 1171 | 297 | 264 | 134 | 101 | 72 | 28 | 38 | 37 | 36 | 26 | 24 | 11 | 15 | 14 | 13 | 10 | 10 | 9 | 7 | 3 | 4 | 3 | 3 | 3 | 3 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TGTGGTCCAGACTAGTTTA................................................................................................................................................................................................................................. | 19 | 2 | 2 | 3.50 | 7 | 1 | 2 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TGTGGTACAGACTAGTTTT................................................................................................................................................................................................................................. | 19 | 2 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TGTGGTACAGACTATTTTA................................................................................................................................................................................................................................. | 19 | 2 | 3 | 2.00 | 6 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AAAGAGTAGTAGTAGGTC............................................................................................................................................................................. | 18 | 2 | 19 | 0.74 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 4 | 1 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 1 |
| .......TGTGGTACAGACTAGTTTTC................................................................................................................................................................................................................................ | 20 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TTGTGGTCCAGACTAGTTT.................................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......ATGTGGTACAGACTAGTTTA................................................................................................................................................................................................................................. | 20 | 3 | 10 | 0.40 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TTTTCTTTGTCGTTGACTAT.................................................................................................................................................... | 20 | 2 | 11 | 0.27 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 |
| .......TGTGGTCCAGACTAGTTTAC................................................................................................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............CAGCCTATTTTTTTTTCTG.......................................................................................................................................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TGTGGTACAGACTAATTT.................................................................................................................................................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TATATTTTCTTTGTCGTTGACTT..................................................................................................................................................... | 23 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......TGTGGTACAGACTATTTTAC................................................................................................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TTGTGGTCCAGACTAGTTTA................................................................................................................................................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................AGGAAGTGCAAAAATGAGAT............................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......TTGTGGTGCAGACTAGTTT.................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TGTGGTACAGACTAATTTA................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TTGTGGTACAGACTAGTTTT................................................................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:17381397-17381647 + | dvi_12362 | GTT--------TTGTACACCAGGTCTGATAAAAAAAAAGGCGGTTGCCAAAGTCACAGGTGAGCAGAGTTTATCATCA---------TCATCAAG------------AAAAA--AAAAGAAACAACAACTGAAAGTCGTTAATTTTTGTTGATATGGATATTTT--TGTATGCCTGGTTTTGACAGACTTC--CTTT------------CCGCTT---TCTACTTTCGTT--TGTC--------AATT-----T-TGA-C-------------------AATTTTTTGCTT--------CACGT--TTTTATTTTATTTGGTTGTTATGCAAAAAAAAAAAAGGGAG |
| droMoj3 | scaffold_6500:22431029-22431223 + | ATA-----------TACAGCAGCTCTGATACGAAAAAAGGCGGCTGCTAAAACAACAGG----------------------------------------------------------------------TGAGAGA-----GCTGTTTGTCATGAAAACGTTTTTCTGT-TGCCTGATTTTGACATACTTC--AGTT-----ACGCTTATCGCTT---GCCATTTTCGGT------------------------------------TGTGAACTTTTTAG-A-TTTTCTTTTTTTTTTTTACGT------------TTTTATCGTTATGCGAAATG-AAAA----AG | |
| droGri2 | scaffold_15252:14816401-14816667 + | GTAACTGTCTTCTTTAAAGCAGGTCTCATAAGAAAAAAGGCAGCTGCCAAATTATCGAGTGAACTG-------CA-CA---------GCACGAAA------------AAAAAGAAAAGTAAGTGAAAAGTGAAA-------AGTGTTTTTGATTTGGATATTTT-----GCGCCTGATTTTGACAGACTTGGCAAATCCGCTTTTCTTTTCGCTT---T-------CGTTTTTATC--------AATT-----T-TGA-CATCGATTTTGAGTTTGGTTT-A-TTTTCCTT--------CATTTTTTTATTCTTCATTTTATCGTTATGCAAGAGAGAAAG----AA | |
| droWil2 | scf2_1100000004521:9995520-9995723 + | ---------------------GGCCTTTTGGGTAAAAAGGCTGCTGCCAAAGCGGCGG---------------CATCATCGGCGACGTCGTCTTCATCGTCATCTGGAG--G--CGGCGGCGGAACAGGTGAGAATTTTCCGTTTTT----------------------------------------------------GCTCTTTCTCTCTCTT---T-------GGTTTATT------TTTGA---GTTTTTGTTGCCATTTTCTTTGCGTTTATTAA-GTCTTTCTT------------TCTCTTTTTTTTCACATCGTCTCCAT------------------- | |
| dp5 | 4_group3:5815624-5815795 - | -------------------CAGGCCTCCTGGGAAAGAAGGCCGCCGCCAAGACATC---------------ATCATCG---------TCATCAGCATCGT-------------------------CAGGTGAGAGA-------------------------------------------------------------CCGCCCTTCCTATGTTTCTCCG-------GGTTTTTC------TTTGA---ATTGTT-TTG-CGTTTTTCCTGAGTTTGTCAAAGTTTTTCATC--------AACTGCTTTCCTTTTCATTTTGTCGTTA-------------------- | |
| droPer2 | scaffold_1:7304109-7304280 - | -------------------CAGGCCTCCTGGGAAAGAAGGCCGCCGCCAAGACATC---------------ATCATCG---------TCATCAGCATCGT-------------------------CAGGTGAGAGA-------------------------------------------------------------CCGCCCTTCCTATGTTTCTCCG-------GGTTTTTC------TTTGA---ATTGTT-TTG-CGTTTTTCCTGAGTTTGTCAAAGTTTTTCATC--------AACTGCTTTCCTTTTCATTTTGTCGTTA-------------------- | |
| droAna3 | scaffold_12916:13097133-13097176 - | -------------------CAGGCCTTCTGAGAAAGAAGGCAGTTGCTAAGCTAACAGG----------------------------------------------------------------------TGAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396580:424538-424581 + | -------------------CAGGCCTTCTGAGAAAGAAGGCGGTTGCTAAGCTAACAGG----------------------------------------------------------------------TGAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000490751:1105876-1105898 - | GT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTGTTTTATTTGGTTGTTA-------------------- | |
| droEug1 | scf7180000409122:683057-683136 + | TTT-------------------CCCCTTTTTGA----------------------------------------------------------------------------------------------------------------------------GTTTC-------------------------------------------------------------CTTCGCT--TGCTAGTAGTTGA---CA-------------------------------------------------TGTTGCTTTCATTTTCATTTTGTCATAA---------AAAAATGTGTG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/16/2015 at 11:19 PM