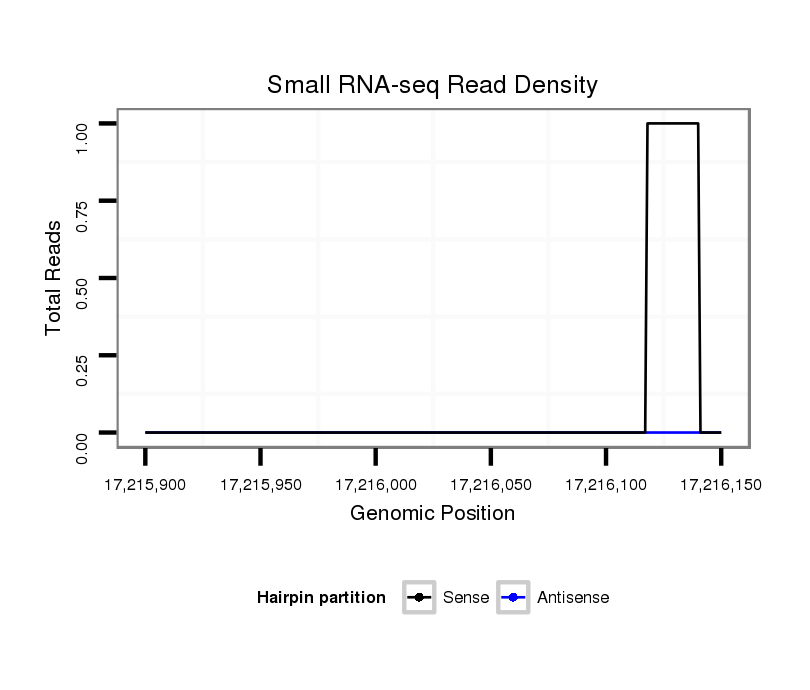

ID:dvi_12341 |
Coordinate:scaffold_12963:17215950-17216100 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ17713-cds]; exon [dvir_GLEANR_2284:2]; intron [Dvir\GJ17713-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTTAATTTTCGGCATGCTGTTGCGTTCCTTCCGGTTCTTTTCCTGTTTTTAGCTATAATTTATCATAGATTTTTTGCGACTCGCGCTAGCCACGTTTTTCGATGTTTGGGCTATTAAACAAATGAGCTGCACATTTAGCCATTCCAGCAAGTTGGCCATTTATCAGTGTCAGCACGTGGCAACGCTGAAATTCCACGACAGGATTCAGTTGCGCCATGTCCTCGCTAACGGTTACACTGCGCGTGCTGGCC **************************************************.((...((((((...))))))....))...(((((...(((((((.......(((((((....)))))))((((.((((......)))))))).......((((((.(((....))))))))))))))))..))).)).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
V053 head |
SRR1106729 mixed whole adult body |
SRR060673 9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................TCCTCGCTAACGGTTACACTGCG.......... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................TAGCAGTGTCATTACGTGGC....................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................CTCCCTAACGGTGACAGTGCG.......... | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................CAGGATTCAGTTGGGC..................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................CGCGCCATCGCTGAAATTCC......................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................GATGTGTGGGCTATGAAAAA................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 |
|
AAATTAAAAGCCGTACGACAACGCAAGGAAGGCCAAGAAAAGGACAAAAATCGATATTAAATAGTATCTAAAAAACGCTGAGCGCGATCGGTGCAAAAAGCTACAAACCCGATAATTTGTTTACTCGACGTGTAAATCGGTAAGGTCGTTCAACCGGTAAATAGTCACAGTCGTGCACCGTTGCGACTTTAAGGTGCTGTCCTAAGTCAACGCGGTACAGGAGCGATTGCCAATGTGACGCGCACGACCGG
**************************************************.((...((((((...))))))....))...(((((...(((((((.......(((((((....)))))))((((.((((......)))))))).......((((((.(((....))))))))))))))))..))).)).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060658 140_ovaries_total |
SRR060664 9_males_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................AGGCGGTCCAGAAGCGATTGC..................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GCGGTTGCCGGTGTGACGCGC........ | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................GTCGTCCACCATTGCGACATT............................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................AAGGCCAAGGAAAGGACAT............................................................................................................................................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................CGGTCCAGAAGCGATTGC..................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ACCCGACAATTTGCTAACTC............................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................CGCAGGGAAGGCCAGGAGA................................................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................GGTCCTGTCCTAAGTATAC........................................ | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:17215900-17216150 + | dvi_12341 | TTTAATTTTCGGCATGCTGTTGCGTTCCTTCCGGTTCTTTTCCTGT-TTTTAGCTATAATTTATCATAGATT---TTTTGCGA--CTCGCGC----------------------TAGCCACGTTTTTCGATGTTTGGGCTATTAAACAAATGAGCTGCACATTTAGCCATTCCAGCAAGTTGGCCATTTATCAGTGTCAGCAC-GTGGCA--ACGCTGAA--------ATTCCACGACAGGA----------------------------TT---CAGTTGCGCCATGTCCTCGCTAACGGTTACACTGCGCGTGCTGGCC |
| droMoj3 | scaffold_6500:22256620-22256838 + | TT------------------------------------TTTCCCATCTCTTGACTATAATTTATCATCGATTT--TTTTGCGA-ACTTGTTC----------------------TGGCCGCATTTTCTGATGTCTGGTCTATTAAAGAAATGAGCTAAACATC-ATTATTTCCGGCCGGCCGGCCATTTGTCACTGTCAGCAC-GTGGCGTCGCACTCGA--------ACCGGGCAACAGGA----------------------------AT---CAGTTGAACCATGTCCGTGCTGACGGTTGCATTGCGCGTGCTCGCG | |
| droGri2 | scaffold_15252:14642366-14642596 - | ---------------------------------------------------------AATTTATCAAAAATTGTTTTTGGCGACACTTG--CTCCTCTTTCTCGTTGTCATTTTTGG----------------CTACACTATTAAACAAATGAGCTGAACATTT-------------AGTTGGCCATTAATCAGTGTCGGCATTGTGGCA--GCAATCGACCAACAACACCACCCACCAGCAACCAAAAGTTGCATGGCGAGTAAATGGTTTTAGTTCTTGCACCATGTCCTTGCTAACGGTAACGCTCCGCGTGTTGGAT | |
| dp5 | 4_group3:10260105-10260157 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGAGA----------------------------GC---CAGTCTTACCATGTCCTGCCTAACGAGAACACTGCAGCTGTTGCAA | |
| droPer2 | scaffold_8:1470208-1470260 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGAGA----------------------------GC---CAGTCTTACCATGTCCTGCCTAACGAGAACACTGCAGCTGTTGCAA | |
| droKik1 | scf7180000302684:287051-287095 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTTGCGCCATGTCCTGTCTAACTGGCATTTTGCAGCTGCTGCAA | |
| droFic1 | scf7180000454043:187235-187285 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGA----------------------------GT---CAGTTGCGCCATGTCCTGTCTAACGAGCATTCTGCAGCTGCTGCAA | |
| droEle1 | scf7180000491338:2074355-2074405 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGA----------------------------GT---CAGTTGCGCCATGTCCTGTCTAACGAGCATTCTGCAGCTGCTGCAA | |
| droBia1 | scf7180000302422:5583379-5583431 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGGGG----------------------------GT---CAGTTGCGCCATGTCCTGTCTAACGAGCGTTCTGCAGCTGCTGCAA | |
| droTak1 | scf7180000415867:341968-342019 - | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGA----------------------------GT---CAGTTGCGCCATGTCCTGTCTAACGAGCTTTCTGCAGCTGCTGCAA | |
| droEug1 | scf7180000409447:63887-63936 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GA----------------------------GT---CAGTTGCACCATGTCCTGTCTAACGAGCATTCTGCAGCTGTTGCAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/16/2015 at 10:29 PM