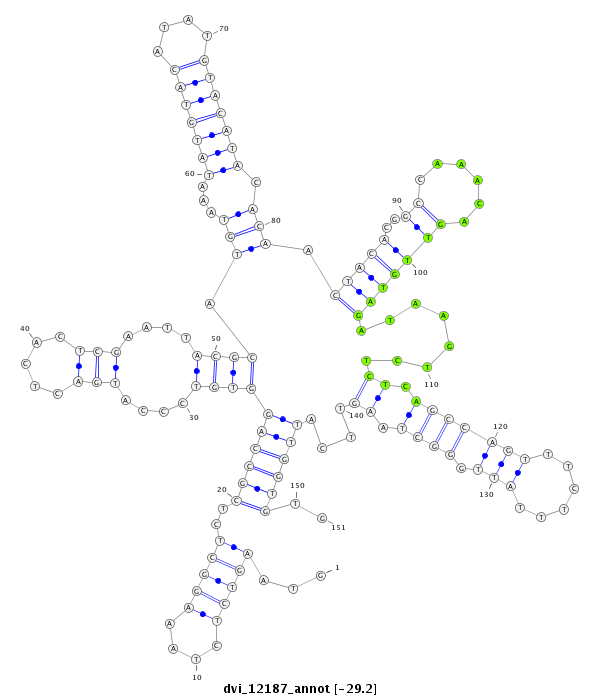
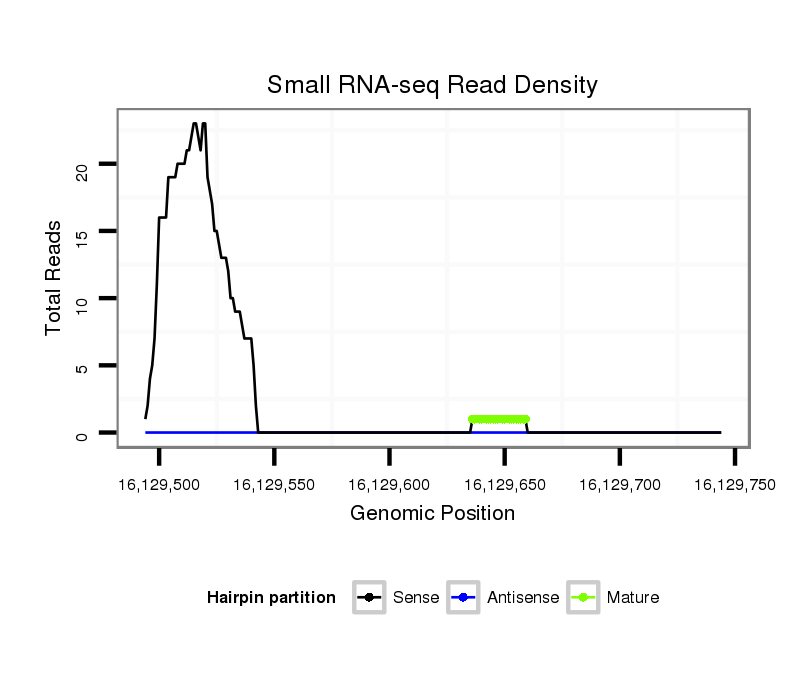
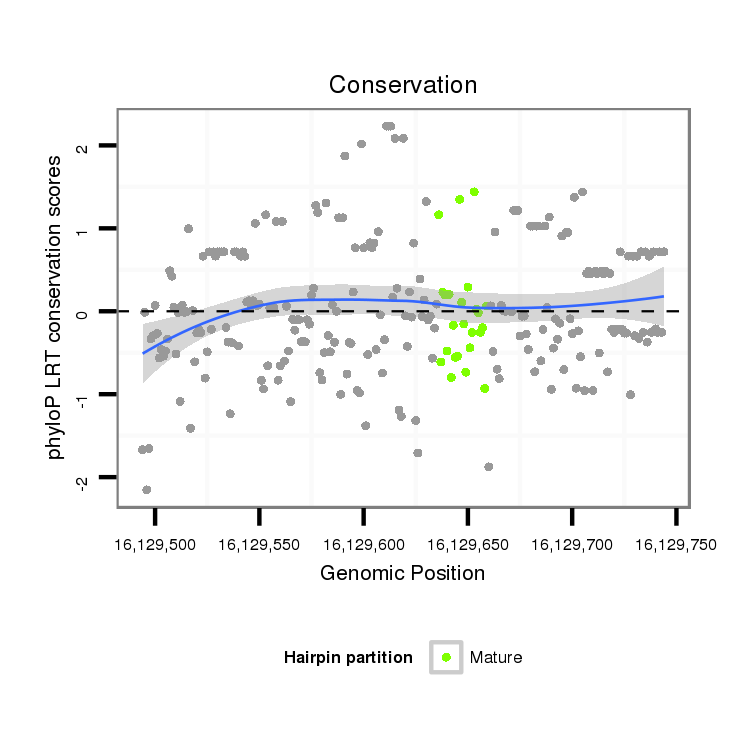
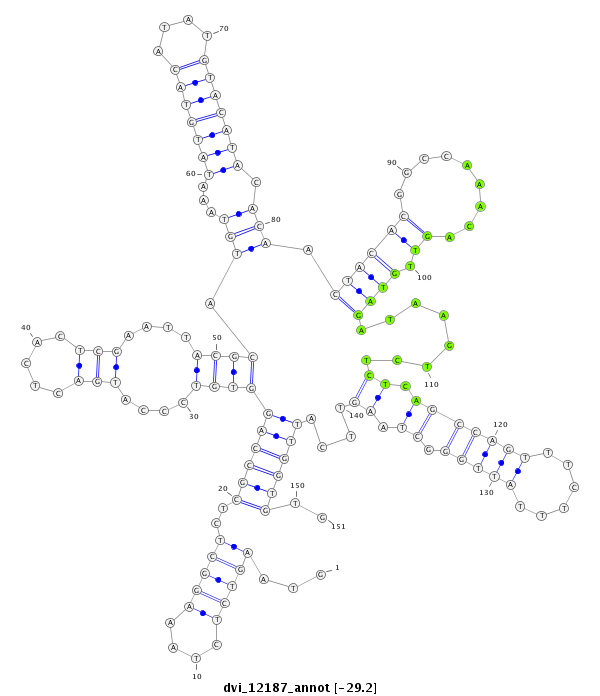
ID:dvi_12187 |
Coordinate:scaffold_12963:16129544-16129694 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -29.2 | -29.2 | -29.2 |
 |
 |
 |
exon [dvir_GLEANR_2245:2]; CDS [Dvir\GJ17670-cds]; intron [Dvir\GJ17670-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CACGCCCACCATCTGCGGCGAGGGCGTAACGGAGTGTTTTGCCAAATCAGGTAAGTCTCTAAAGGCTCTCGCCAGGTGTCCCATGACTCACTCGAATTACGCATGTAAATATGTACATATGTACATACACAACTACACGGCCAAACAGTTGTAGATAAGTCTCTCAGCCAGTTTCTTTATTGGGCTAAGTTCATTGGTGTGCCTATCTCCGGGCAGCTGAACACGGTGCTCAAGAGCCGATAGCAATAAAT **************************************************...(((((....)))))..((((((((((....(((.....)))....)))).(((...(((((((....))))))).))).(((((..((......)))))))........((.(((((((......))).)))).))....))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060665 9_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
M061 embryo |
SRR060664 9_males_carcasses_total |
SRR060667 160_females_carcasses_total |
M027 male body |
SRR1106729 mixed whole adult body |
V047 embryo |
SRR060662 9x160_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060659 Argentina_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060678 9x140_testes_total |
SRR1106721 embryo_10-12h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................GTAACGGAGTGTTTTGCCAAAT............................................................................................................................................................................................................ | 22 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GTAACGGAGTGTTTTGCCAAATC........................................................................................................................................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GTAACGGAGTGTTTTGCCAAATCA.......................................................................................................................................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......CACCATCTGCGGCGAGGGCGT................................................................................................................................................................................................................................ | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....CCACCATCTGCGGCGAGGGC.................................................................................................................................................................................................................................. | 20 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CACGCCCACCATCTGCGGC........................................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...GCTCACCATCTGCGGCGAGGGCGT................................................................................................................................................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AGGGCGTAACGGAGTGTTTTGC................................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........ATCTGCGGCGAGGGCGTAACGGAGTGT...................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....TCACCATCTGCGGCGAGGGC.................................................................................................................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CGGAGTGTTTTTCCAAATCA.......................................................................................................................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CGCCCACCATCTGCGGCGAGGGC.................................................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...GCCCACCATCTGCGGCGAGG.................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................AACGGAGTGTTTTGCCAAATCAGATGC..................................................................................................................................................................................................... | 27 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................AAAGGAGTGTTTTGCCAAATC........................................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GTAACGGAGTGTTTTGCC................................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................GAGGGCGTAACGGAGTGT...................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......CACCATCTGCGGCGAGGGC.................................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................AAACAGTTGTAGATAAGTCTCTCA..................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................CGAGGGCGTAACGGAGTG....................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................GGGCGTAACGGAGTGTTT.................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........ATCTGCGGCGAGGGCGTAAC............................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......CACCATCTGCGGCGAGGGCGTAAC............................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGTGAATCTCTAAAGGCTGTC..................................................................................................................................................................................... | 21 | 3 | 2 | 1.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........ATCTGCGGCGAGGGCGTAACGGA.......................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....CCACCATCTGCGGCGAGGGCGTAA.............................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................GAGTGTTTTGCCAAATCAGATGC..................................................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......CACCATCTGCGGCGAGGGCGTA............................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............GCGGCGAGGGCGTAACGG........................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....CCACCATCTGCGGCGAGGGCGT................................................................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CCCACCATCTGCGGCGAGGGCGTA............................................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CGCCCACCATCTGCGGCGAGGG................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................ACGGAGTGTTTTGCCAAATC........................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........ATCTGCGGCGAGGGCGTAAA............................................................................................................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .ACGCCCACCATCTGCGGCGAGGGCGT................................................................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CCCACCATCTGCGGCGAGGGC.................................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGTGAATCTCTAAAGGCTGTCG.................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................GGTGAATCTCTAAAGGCT........................................................................................................................................................................................ | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................TGTCTCCGGGAAGCTGAACA............................ | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TTGGTGGCCCTATCTCCGT....................................... | 19 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................CAATTTCTTTATTGGGCTTA............................................................... | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................GGGCGTAACGGAAGGTTTC................................................................................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................CACGGTGTTCTAGAGCC............. | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................................ACGAAGCTGAACACGGTGCT..................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTGCGGGTGGTAGACGCCGCTCCCGCATTGCCTCACAAAACGGTTTAGTCCATTCAGAGATTTCCGAGAGCGGTCCACAGGGTACTGAGTGAGCTTAATGCGTACATTTATACATGTATACATGTATGTGTTGATGTGCCGGTTTGTCAACATCTATTCAGAGAGTCGGTCAAAGAAATAACCCGATTCAAGTAACCACACGGATAGAGGCCCGTCGACTTGTGCCACGAGTTCTCGGCTATCGTTATTTA
**************************************************...(((((....)))))..((((((((((....(((.....)))....)))).(((...(((((((....))))))).))).(((((..((......)))))))........((.(((((((......))).)))).))....))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
V053 head |
SRR1106729 mixed whole adult body |
SRR060666 160_males_carcasses_total |
SRR060660 Argentina_ovaries_total |
SRR060672 9x160_females_carcasses_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................GGTCCACATTGTACTGAGCGAG.............................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TCAAGAAACCACACGGCAAGA........................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ACATGTAAGTGTTAATGGGC................................................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TGTGTAGGTTGATGTGCCGGT............................................................................................................ | 21 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................TCGTCCATACCGAGATTTC........................................................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................ACGGTTTAGTTCATTCAA.................................................................................................................................................................................................. | 18 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TGTTTGTATTGGTGTGCCGGT............................................................................................................ | 21 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................GGACTTGAGCCATGAGTTC................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................TGCTGTGCCGGTTAGCCAA..................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................TGCATGTATACATGTATGTG......................................................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:16129494-16129744 + | dvi_12187 | CACGCCCA-CCATCTGCGGCGAGGGCGTAACGGAGTGTTTTGCCAAATCAGGTAAGTCT------------------------------------------------------------------------------------CTAAAGGCTCTCGC---------CAGGTGTCCCATGACTCACTCGAATTACGCATGTAAATATGTACATATGTACATACACAACTACAC-GGCCAAACAGTTGTAG-------------------------------ATAAGTCTCTCAGCCAGTTTCTTTATTGGGCTA--AGTTCA-TTGGT--------------GTGCCTATCTC-CGGGCAGCTGAACACG-----------------------GTGCTCAAGAGCCGATAGCAATAAAT |
| droMoj3 | scaffold_6500:5589865-5590118 + | CACATCCC-AAACCTGTACAGATGCTCAAGCCGAATGTTTTGTGAAAACAGGTGAGATACACTCGTTGGGACCCATGAATCATCCACAATTGGGATTGGCGAACACACA--------------------------------------------------------------------------------------------------GCTGTAT-------------------------ATGGTTACAGATAAGTTAATAGTTGGCT-----------------------TACCAAAGTTCTTTATT----------------GGACACAGCTTACTCGGAGCACTTATCTG-CAGCCAGGTGGACAAGCGATAAGAAATGTATTTTCATATACATTTTTGCGCCTACAACGATGAGT | |
| droGri2 | scaffold_15252:13555651-13555853 + | CAATCCCA-CCGCATGTCCGGCTGATGTTACCCAGTGTTTTATTCAAACAGGTAAGTCGCCTCCGAT-----------------TTTGGTTTAGGTTAGGTGACTCACAAGTATATATGCCAATAGATAAGTAGGACAAGTTCATTTTAGTACTTATCGG-------------------------------TGTGTGCATGGGTGTGTCTGTATCTGTGTGTGTGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCTCAAGTGACGATAACAACAAAT | |
| droWil2 | scf2_1100000004762:1090390-1090501 + | CTGTAACT--GCTATATACGAATGTT----------------------------------------------------------------------------------------------------------------------------------------------------CAATGTTTCTTTT----TTTTTATGTATGTATATATGTATGTATGTACATAG---------------------------ATTTTCATTTGGCTTAAATTGTTGCAGTTGCTTCTC--------------------------------------------------------------A-CAG-------------------------------------------------------------- | |
| droAna3 | scaffold_13137:1024666-1024742 - | G--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACATACATATGTATGTATGTATATACAAAAATACAA-ATGCATACCGCCCTAG-------------------------------TGAAATTAGCCGGTCATAT--------------------------------------------------TTTT----------------------------------------------------------------- | |
| droBip1 | scf7180000396740:263627-263742 - | AAT-TATACATATAAATA-------------------------------------------------------------------------------------CATATA----------------------------------------------------------------------------------TATGTATGTATGTATATGGATATGTAAGTATGTA-----------------------GATATTTTCGTATTTAGCT-----------------------TGCT----------ATTGAAAGACCATTTCT-TTGCCGTGGCTTT--------------CTC-TGG-------------------------------------------------------------- | |
| droKik1 | scf7180000302345:21871-21971 - | CAGATA-------------------------------------------------------------------------------------------------------------------------------AAACAAGTTTCTTAAAGGACCCAGCAGCCGGCAAAGGAGACTCGTGACACGC---ACTTATGTACATACATATGTACATATGTATGTACATATGTACA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453809:194322-194413 - | TTTGCCAA-TTGAGAAA----------------------------------AAATGTCG------------------------------------------------------------------------------------ATAAAAG-TCGCGC---------AGAATTTTTCA-----CAAATAAATTGAGTATGTATGTATATACATATGTATATATATAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEle1 | scf7180000491011:433451-433523 + | T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACATACATATGTATGTACATATGTACTTGCACATTTGTA-----CATACAAA----------------TCTGGTTCAAG---------AAATGT---------------------------------------------------------------CTCTGAG-------------------------------------------------------------- | |
| droRho1 | scf7180000778085:87180-87294 + | GAGTTATCTTTGTAAATA-------------------------------------------------------------------------------------CATACA--------------------------------------------------------------------------------------------------TATGTATATGCTTACATACATA---------------CACACATACGCCATTGTTTGACTCAGA---------------------------CCTCCAATTTGGCTA--ACTTGATTTGAT--------------TTGCATAGTTCTTGG-------------------------------------------------------------- | |
| droBia1 | scf7180000301454:29997-30058 - | AGT-TGTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------CCATGCGGCAAACAATTTGCACACATATAAATATACATATGTACTTATATAAGTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000413976:13069-13136 - | GAAGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTATGTACGTACGTACATATGTAAGTGCATAAACTTAACGAGTACGCAGGTATAC-------------------------------ATATGT------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/19/2015 at 05:25 PM