
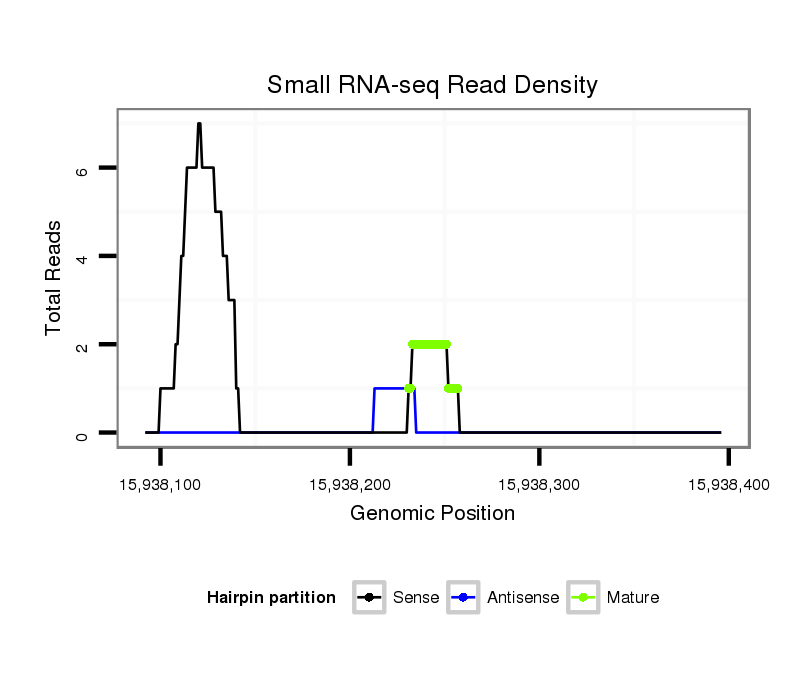
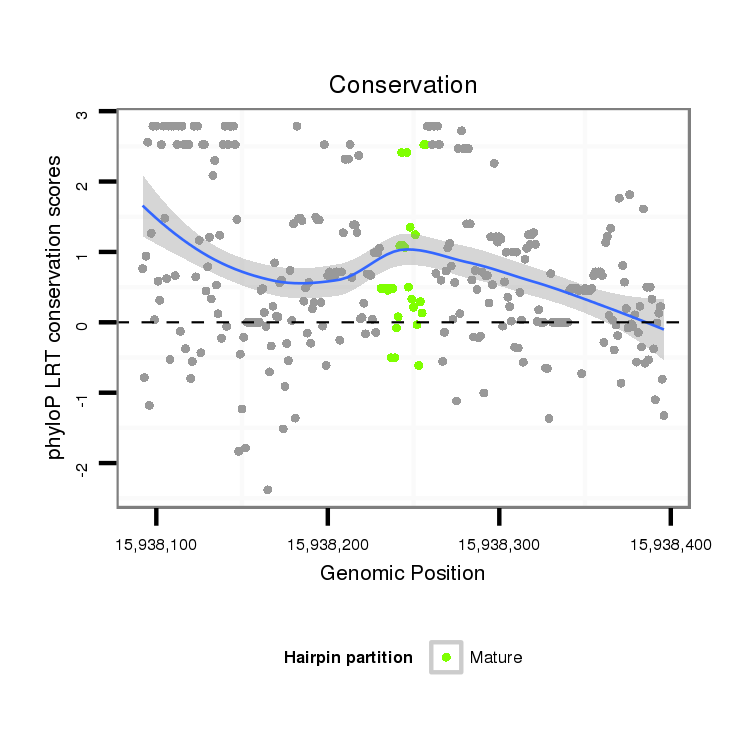
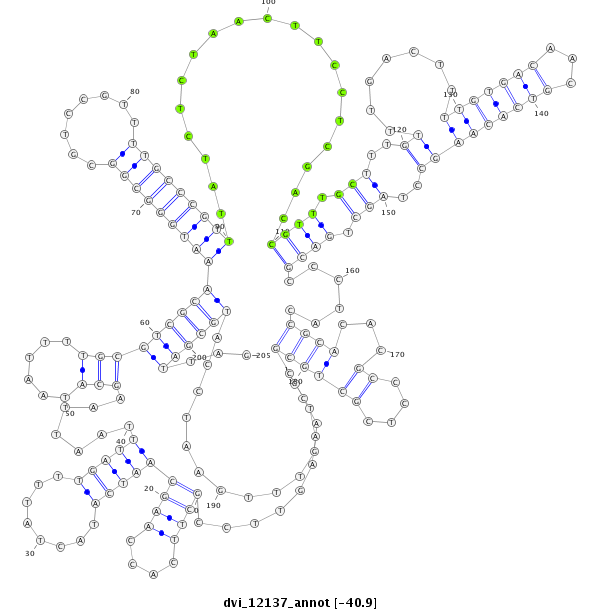
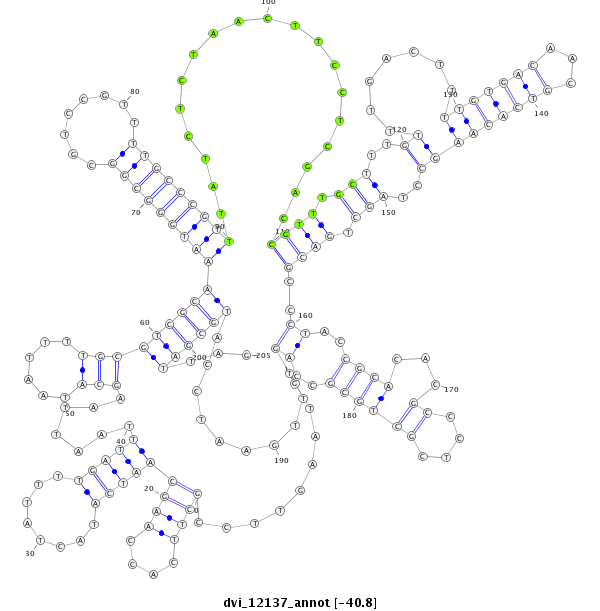
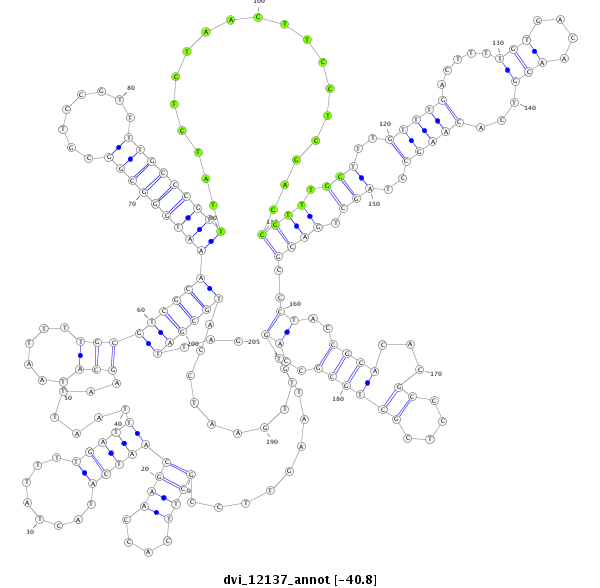
ID:dvi_12137 |
Coordinate:scaffold_12963:15938142-15938346 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -40.9 | -40.8 | -40.8 |
 |
 |
 |
CDS [Dvir\GJ17655-cds]; exon [dvir_GLEANR_2231:2]; intron [Dvir\GJ17655-in]; exon [dvir_GLEANR_2231:1]; CDS [Dvir\GJ17655-cds]; intron [Dvir\GJ17655-in]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------#################--------------------------------- AGCATGAAAATGATTTGTTCGTTTCGGCAATGACCCCGAGCGAAGTGCATGTAAGTTCCGCTTCACCAAGCAATCATACTATTTTGATTTAATTAAGCATAATTTTGCGTCGCAAATGGGCGGCGTCCGTTTTGCCCGTTTATCTCTAACTTCCTCGACCGTTTGCTTTGTTTGACTTTTGTGACAACGTCACAAGCCTAGCTGACGCCCTACCGCACACGCCCTCGCTGCGCCAGTTTGAATCCATGCGATTAGGCGACGGACTTATCACAGTGCACATTCATTTATTATCTTTATCAATCTCG **************************************************.........((((....))))(((((........))))).......(((......)))(((((((((((((((........)))))))))...................((((.(((..(((((.....(((....)))...)))))..))).))))......((((...((....))))))..............))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060676 9xArg_ovaries_total |
SRR060657 140_testes_total |
SRR060659 Argentina_testes_total |
SRR060664 9_males_carcasses_total |
SRR060665 9_females_carcasses_total |
SRR060666 160_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060681 Argx9_testes_total |
V047 embryo |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................CGTTTCGGCAAAGACCCCGAG......................................................................................................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................AATGACCCCGAGCGAAGTGCAT............................................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................TCGTTTCGGCAATGACCCCGAGCGAA..................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................GTTCGTTTCGGCAATGACCCC............................................................................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................TTATCTCTAACTTCCTCGACCGTTTGC........................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................TTCGGCAATGACCCCGAGCGAAGTGC................................................................................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................ATCTCTAACTTCCTCGACC................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................CGTTTCGGCAATGACCCCGAGC........................................................................................................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........AATGATTTGTTCGTTTCGGCAA................................................................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................TTTCGGCAATGACCCCGAGCGAAGTGC................................................................................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GCCCGTCTATCTCTGACT.......................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................AAGCCTAGCGGAGACCCTA............................................................................................. | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................ATGCGATGAGAGGACGGA.......................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................CCGTATGTTTTTTTTGACTTT.............................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TCGTACTTTTACTAAACAAGCAAAGCCGTTACTGGGGCTCGCTTCACGTACATTCAAGGCGAAGTGGTTCGTTAGTATGATAAAACTAAATTAATTCGTATTAAAACGCAGCGTTTACCCGCCGCAGGCAAAACGGGCAAATAGAGATTGAAGGAGCTGGCAAACGAAACAAACTGAAAACACTGTTGCAGTGTTCGGATCGACTGCGGGATGGCGTGTGCGGGAGCGACGCGGTCAAACTTAGGTACGCTAATCCGCTGCCTGAATAGTGTCACGTGTAAGTAAATAATAGAAATAGTTAGAGC
**************************************************.........((((....))))(((((........))))).......(((......)))(((((((((((((((........)))))))))...................((((.(((..(((((.....(((....)))...)))))..))).))))......((((...((....))))))..............))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
V047 embryo |
M027 male body |
M047 female body |
V053 head |
SRR060655 9x160_testes_total |
V116 male body |
SRR060689 160x9_testes_total |
SRR060674 9x140_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
M061 embryo |
SRR1106729 mixed whole adult body |
SRR060679 140x9_testes_total |
SRR060681 Argx9_testes_total |
SRR060662 9x160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................CGGTGGGATGGCGTGTGCGGGA................................................................................ | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CCGCAGGCAAAACGGGCAAATA.................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................GGAACGCTAATCCGCTGGC........................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GAGCTTGAAGGTGCTGGAA............................................................................................................................................... | 19 | 3 | 5 | 0.60 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CTACACGGTCTAACTTAGGTA.......................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GCAGCCGTGTGCGGGAGCGA............................................................................ | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TTCAGGGCCAAGTGGTTC........................................................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AGCTTGAAGGTGCTGGAA............................................................................................................................................... | 18 | 3 | 20 | 0.30 | 6 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................GAGCTGCCAAACGAAACCGAC................................................................................................................................... | 21 | 3 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ................................TGGGGCACGCTGAACGTACA............................................................................................................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TTTCAGGGCCAAGTGGTTC........................................................................................................................................................................................................................................... | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................CTGGCCAACGAACCAAACTC................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................CGGGCAAGAAGAGATTG........................................................................................................................................................... | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................GGAGCTGGCAAGCGAATCT...................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................ATTCAGGGCCAAGCGGTTC........................................................................................................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CCGCCGCAGGCTGAAAGGG........................................................................................................................................................................ | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................TAGTGGCACGTGTAGGTGA.................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TCGTACTTTCACGAAACTAG............................................................................................................................................................................................................................................................................................. | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TTTCAAGGCCAAGCGGTTC........................................................................................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................GCAGTGTTCGCATGGCCT.................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................GCGCTTCACGCACAGTCAA........................................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .CGTAATGTTACTAAACAA.............................................................................................................................................................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:15938092-15938396 + | dvi_12137 | A---GCATGAAAA---------TGATTTGTTCGTTTCGGCAATGACCCCGAGC---------GAAGTGCATGTAAGTTCCGCTTCACCAAGCAATC--------------------A---TAC-T-------------------AT-T-T--TGATTTAATTAAGCATAATT-TT--GCGTCGCAAAT-GGGCGGCGTCCGTT--------TTGCCCGT-TTATCTC-TA---ACTTCCTC---------------GAC-----CGTTTGCTTTGTTTGACTTT---------------T---G-TGACAACGTCACA-AG-CCTAGC---TGACGCCCTAC-----CGCACACGCCCTCGCTGCGCCAGTTTGAATCCATGCGATTAGGC-GACG-----GAC-----TTATCACAGT-------------------------GCACAT----TCATTTATTA-------------------------T-------CTTTAT------CAATCT------CG |
| droMoj3 | scaffold_6500:5382358-5382644 + | AGC---AAGAAAA---------TGATTTATTCGTATCAGCAATGACGCCGAGC---------GAAGTGCATGTAAGCAAGCA--------GCGAAC---------------------AAATAT-T-------------------GT-A-T--TTATTCAATTAAGCATAAATTTGATGGGTCGCAAGTGGGGCGGCGTCCGAG------------CTGTTTTATCTTCTGCTTA------CAA----------------TGCT-CGCGTGCTTTGTTTGA-TT----------------T---G-TGACAACGTCACAGAG-TCTCGC---TGACGCCCCTCTAGCC------------------------------------GATTAGGAAGACGACTGCGAC-----TTATCGCAGTGCAGTTCATCTCTAGT-GTAAGCTAA--T--TA---------------------T-----------------------ATATATA------TATAT------AT | |
| droGri2 | scaffold_15252:13366672-13366969 + | AGC---ATGAAAA---------TGATTTATTCGTATCCGCAATGACGCCCAGT---------GAGGTGCATGTAAGTTACAC--------------------------------------------------------ATTTATTT-CAA--TTATTAAATTAACTTGAATT-AT--GCGTCGCAAAT-GGGCGGCGTCTGTC----CT------------------------TTATTATTAGCTTTAT---CTTTGTGTTTTACTGTTGCTTTGTGTGT-GTG---------------T---G-TGTCAACGTCACA-ATGCTTTGCGAGGGGCGGCAACA-----ACA---AATTTTTGTTGAT---TAGA------------------------------CTTATTTTATCGCAT---------------CTGCATTGCTTA--T--AATATGTATAA---TTATGCTTGTACATT--------TT-----ACAT------TT----TT---GTTGATAT | |
| droWil2 | scf2_1100000004585:8337110-8337347 - | ATGA---AAATAA---------TGATTTATTCGTATCGGCCATAAGTCCGGGTGACGT------TACACACGTAAGTTCAAA---------CAAGCA------------------CACAAAAG-GATGCGTTGGCAGCATTTGCAT-T-T--ACACGCAATTCATACTAATC-GTACG-----------G---GT---CGGG---------------------------------------------------------CGTTGCTCTGGCTTTGTGTGG-CCTCAGCCGCCCCCCTTTT---C-ATGCTACGCCGCT-GC----------TAC-ACTCCCC-----CTC---AAAGCTTATCAGC---G------------------------------------------------T-------------------------TCCAAT----TGAATTT-------------------------------------------------AAACT------CT | |
| dp5 | 4_group3:921274-921541 + | A---GAACGAAAA---------TGATTTATTCGTATCAGCCATGAGTCCCAGTAGCATCGGGGATGTGCATGTAAGTCAAAT---------CAGA-------------GCGTACGCCCAACAG-G-------------------AT-G-C--CTATTAAATTAGGTTTAATTTTT--G-----------GGGCGGCG---CTC--TCCATTTTGC----------------TTGCTTTATCAA--------CCAGAGACTGTTGC-TGTGCTTTGTTTGG-CGT---------------------TGACAACGTCATT-AG----------AGCCGCCCACC-----AGCACACAGCCATGTTGTT---GTTT---------------------------------------------------------------GTGTGCTTA--T--CA---------------------TCAATTGAAT-------------CGTCTCTT------TCTCT------CT | |
| droPer2 | scaffold_1:2401085-2401352 + | A---GAACGAAAA---------TGATTTATTCGTATCAGCCATGAGTCCCAGTAGCATCGGGGATGTGCATGTAAGTCAAAT---------CAGA-------------GCGTACGCCCAACAG-G-------------------AT-G-C--CTATTAAATTAGGTTTAATTTTT--G-----------GGGCGGCG---CTC--TCCATTTTGC----------------TTGCTTTATCAA--------CCAGAGACTGTTGC-TGTGCTTTGTTTGG-CGT---------------------TGACAACGTCATT-AG----------AGCCGCCCACC-----AGCACACAGCCATGTTGTT---GTTT---------------------------------------------------------------GTGTGCTTA--T--CA---------------------TCAATTGAAT-------------CGTCTCTT------TCTCT------CT | |
| droAna3 | scaffold_12943:2511380-2511628 - | G---AAACGACAC---------CGATCTTTTCGTGTCGGCCATGAGCCCCAGCAGCATTGA------ACATGTAAGTGCGGG---------CCA---------------------CCCAGCAG-GAC------------------G-T-A--AACTTAAATTGGCTCAAATT-TT--G-----------AGGCGG---CGCCTGCTTCA-----TCTGT------------TCTCTTTATTAT------CAATAACGACTGTTGCTTTTGCTTTGTTTGG-AT----------------------CGTCGACGTCATA-AG----------TGCCGCCCAC------------------TGTTGCT---CA--------------------------------------------CAGT-----------------GGA----G-TGTAG----TGAG------CAATGTG--TACATT------CTGT-----GTTC------AT----TA---------GC | |
| droBip1 | scf7180000396728:819537-819790 + | G---GAACGACAC---------CGATCTTTTCGTGTCGGCCATGAGCCCCAGCAGTATGGA------ACATGTAAGTGCGAG---------CCA---------------------CCCATCAG-GAT------------------T-T-AC-CCA-TCAATTGGCTCAAATT-TC--G-----------AGGCGG---CGCTCGCTTCA-----TCTGT------------TTTCTTTATTAT------CACTAACGACTGTTGCTTTTGCTTTGTTTGG-AT----------------------CGGCGACGTCATG-AG----------TGCCGCCCAC------------------TGTTGCT---CA--------------------------------------------CAGT-----------------GGA----G-AGTAC----TGAA------CAATGCG--TATACT------CTGG-----GTTC------AT----TA---GCTGC-TA | |
| droKik1 | scf7180000302472:1790445-1790721 - | G---CAATGAGAGCAGCGATCGGGATCTGTTCGTCTCAGCTTTGAGCCCCAGCAGCATCGGAGATGTGCACGTAAGTAGCAC------------GGTTGGCTGGATGAGCTCAATCACAACAG-G-------------------AT-G-C--TTATTCAATTAGCTCTAATT-TC--G-----------GGGCGG---CTCTCGC----------------------------TCTTTATCAGCTGCCG---CGACGACTGTTGCTTTCGCTTTGTTTGG-CTT---------------TCGGG-CGGCGACGTCACG-AG----------TGCCGCCCCCCAAGCCAGCAGAGAACCTTGTTGCT---CA------------------------------------------------------------------------------------------------------TCCATTTAAGCT--CT-----ACTC------TC----TC---------TC | |
| droFic1 | scf7180000453924:1434149-1434397 + | ATCGGAATGAAAG---------CGACCTATTTGTGTCGGCTCTGAGTCCCTCCAGCATCGGGGATATACATGTAAGTCGCAG---------CAACT--------------------ACAACC-GGA--AGCCACCCAGATCGGCAG-C-A--CCCTTAAATTGCCTCCAATT-GC--G-----------GGGCGT---CGGTC------------------------------GTTTTATCAG---------------------CGCTCGCTTTGTTTGG-CTT---------------T---G-TGACTACGTCACG-AG----------TGCCGCCCACC-----AGA---AAAGCTTGTTGCA---TGCT----------------------------------------------------------------------AA--T--AA-----AT-----------------ATTTATT--------T---GCCTTCTC------CACTCC------CA | |
| droEle1 | scf7180000490728:110812-110993 + | ACAGGAATGAAAG---------TGATCTATTTGTGTCGGCACTAAGTCCCAGCAGTATCGGAGATATACACGTAAGTCGGGG---------GAATATACAG-------GCT-----ATATCC-GCA--TGCAACCTACATTCGGAG-C-C--CTATTTAATTAGCTCCAATT-TC--G-----------GGGCGT---CGCTC------------------------------GCTTTATCAA---------------------CATTCGCTTTGTTTGGGCTT---------------T---G-T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779523:121016-121271 + | ACCGGAACGAAAG---------TGATCTATTTGTGTCGGCATTGAGTCCCAGCAGCATGGGAGATATACACGTAAGTCGGGG---------AAGCATAGCC----------------------GGA--TACAGCCCGCACAAAGAG-C-C--CTACTTAAATAGCTACGATT-TC--G-----------GGGCGT---CGCTC------------------------------GCTTTATCAG---------------------CGCTCGCTTTGTTTGG-CTT---------------T---G-TGGCTACGTCACG-AG----------TGCCGCCCACC-----AGC---ACAGCTTGTTGCT---TTAT---------------------------------------------------------------ATA----T------------------------------CCATTTCATTG--CTGCTTCTTCCTCTCT------CAATCC------CC | |
| droBia1 | scf7180000301468:1061849-1062120 - | ACCGCAACGAAAG---------TGATCTTTTTGTGTCCGCCCTGAGTCCCAGCAGCATCGGCGATGTCCACGTAAGTGGGGT---------CAGTGGACCC-------------ACCTACCCGGGG--AGCAGCCCGCATTCAGAG-C-TCTCTATTGAATTAGCTCCAATT-TC--G-----------GGGCGT---CGGTC------------------------------GCTTTATCAG---------------------CGTACGCTTTGTTTGG-CTT---------------T---G-CAACTACGTCACG-AG----------TGCCGCCCACC-----AGC---GAAGCTTGTTGCT---CCGT---------------------------------------------------------------AGA----ATTAGA--AAATGTATATGTA-------------------------TCTAATGCC------ACTTCCGTTCC------CA | |
| droTak1 | scf7180000415914:647329-647575 - | ACCGAAATGAAAG---------TGATTTATTTGTTTCGGCTTTGAGTCCCAGCAGCATCGGCGATATCCATGTAAGTAGCGC---------CGATAAACCC----------------------G-G--AATAACCCGCATCCGGAGGT-TCTATACTAAATTAGTTCCAATT-TC--G-----------GGGCGT---CGGTC------------------------------GCTTTATCAG---------------------CGTTCGCTTTGTTTGG-CTT---------------T---G-TGGCTACGTCACG-AG----------TGCCGCCCACC-----AGC---AAAGCTTGTTGCT---CCCT---------------------------------------------------------------GGA----AATATAT------------------------CCC-------------------CTTTTCT------CTAATT------TA | |
| droEug1 | scf7180000409554:2638974-2639225 - | ATCGGAATGAAAG---------TGATCTATTTGTGTCGGCACTGAGCCCCAGCAGTATTGGGGATATCCACGTAAGTGAGGG---------CAATAAACCC----------------------GTG--AGCAGCCCGCATCAGGAG-G-T--CTATTAAATTAGCTTCCATT-TC--G-----------GGGCGT---CGGTT------------------------------GCTTTATCAG---------------------AGTTTGCTTTGTTTGG-TTT---------------T---GATGACTACGTCACG-AG----------AGCCGCCCACC-----AGC---AAAGCTTGTTGCT---TCAT---------------------------------------------------------------AGA----AA---------TGAGTTTGAG-------------------------TATGAG---TTTTT------TACACC------CT | |
| dm3 | chr2L:3449916-3450171 + | ATCGCAACGAAAG---------TGATCTCTTCGTGTCGGCCTTGAGTCCCAGCAGCATCGGGGATATACACGTAAGTTGGGG---------CACGAAACAC----------------------GGG--AGCAGCCCGCATCGACAG-C-CC-CCAGTAATTGAGCTCCCGTT-TT--G-----------GGGCGTCGTCGGTC------------------------------GTTTTATCAG---------------------CGCTCGCTTTGTTTGG-TTT---------------T---G-TGGCTACGTCACG-AG----------TGCCGCCCACC-----AGC---AAAGCTTGTTGCT---CCAT---------------------------------------------------------------AGA----AACACAA----TGAATTTGCG-------------------------TCTTAATGC------ATT-------C------CC | |
| droSim2 | 2l:3309602-3309857 + | ATCGCAACGAAAG---------TGATCTTTTCGTGTCGGCCTTGAGTCCCAGCAGCATCGGGGATATACACGTAAGTTGGGT---------CACGAATCGC----------------------GGG--AGCAGCCCGCATCGGCAG-C-CC-CCAGTAATTGAGCTCCCATT-TC--G-----------GGGCGTCGTCGGTC------------------------------GCTTTATCAG---------------------CGCTCGCTTTGTTTGG-TTT---------------T---G-TGGCTACGTCACA-TG----------TGCCGCCCACC-----AGC---AAAGCTTGTTGCT---CCAT---------------------------------------------------------------AGA----AACACAA----TGAATTTGCG-------------------------TCTTAATGC------ACTC-------------CC | |
| droSec2 | scaffold_129:12034-12289 + | ATCGCAACGAAAG---------TGATCTTTTCGTGTCGGCCTTGAGTCCCAGCAGCATCGGGGATATACACGTAAGTTGGGT---------CACGAAACGC----------------------GGG--AGCAGCCCGCATCGGCAG-C-CC-CCAGTAATTGAGCTCCCATT-TC--G-----------GGGCGTCGTCGGTC------------------------------GCTTTATCAG---------------------CGCTCGCTTTGTTTGG-TTT---------------T---G-TGGCTACGTCACA-TG----------TGCCGCCCACC-----AGA---AAATCTTGTTGCT---CCAT---------------------------------------------------------------AGA----AACACAA----TGAATTTGCG-------------------------TCTTAATGC------ACTC-------------CC | |
| droYak3 | 2L:3437721-3437979 + | ATCGGAACGAGAG---------TGATCTTTTCGTGTCGGCCCTGAGTCCCAGCAGCATCGGGGATATACACGTAAGTGGGGG---------CAAAAAACGC----------------------TGA--TGCAGCCCGCATCGGTAG-C-C--CCAGTAATTGAGCTCCTATT-TC--G-----------GGGCGTCGTCGGTC------------------------------GCTTTATCAG---------------------CGCTCGCTTTGTTTGG-TTT---------------A---G-TGGCTACGTCACG-AG----------TGCCGCCCACC-----AGC---AAAGCTTGTTGCT---CCAT---------------------------------------------------------------AGG----GACACAT----AGAATTCGCG-------------------------TCTTATTGC------ACTCCCAAT---------C | |
| droEre2 | scaffold_4929:3482293-3482548 + | ATCGGAACGAAAG---------TGATCTTTTCGTGTCGGCCTTGAGTCCCAGCAGCATCGGGGATATACACGTAAGTGGGGG---------CAAGCATCGC----------------------GGG--AGCAGCCCGCATCGGTAG-C-C--CCAGTAATTGAGCTCCCATT-TC--G-----------GGGCGT---CGGTC------------------------------GCTTTATCAG---------------------CGCTCGCTTTGTTTGG-TTT---------------T---G-TGGCTACGTCACG-AG----------TGCCGCCCACC-----AGC---AAAGCTTGTTGCT---CCAT---------------------------------------------------------------TGG----GACACAT----TGAATTTGCG-------------------------TCTTATTGC------ACTCCCAAT---------C |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/19/2015 at 05:20 PM