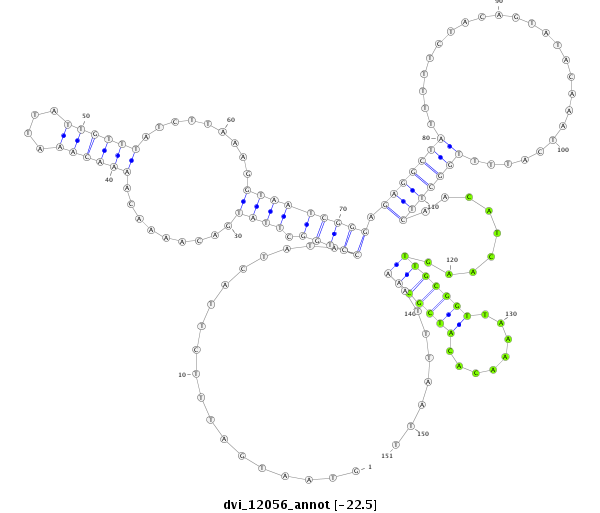
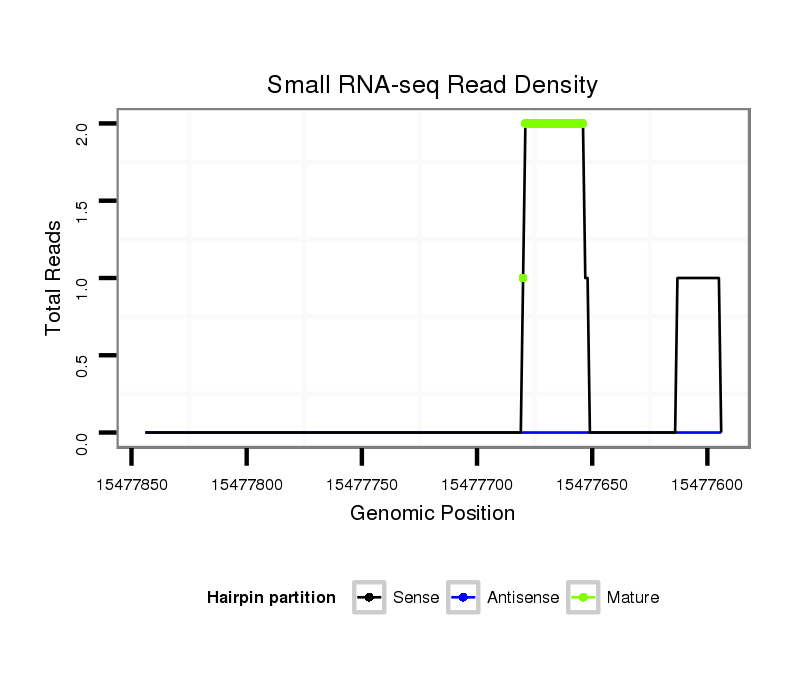
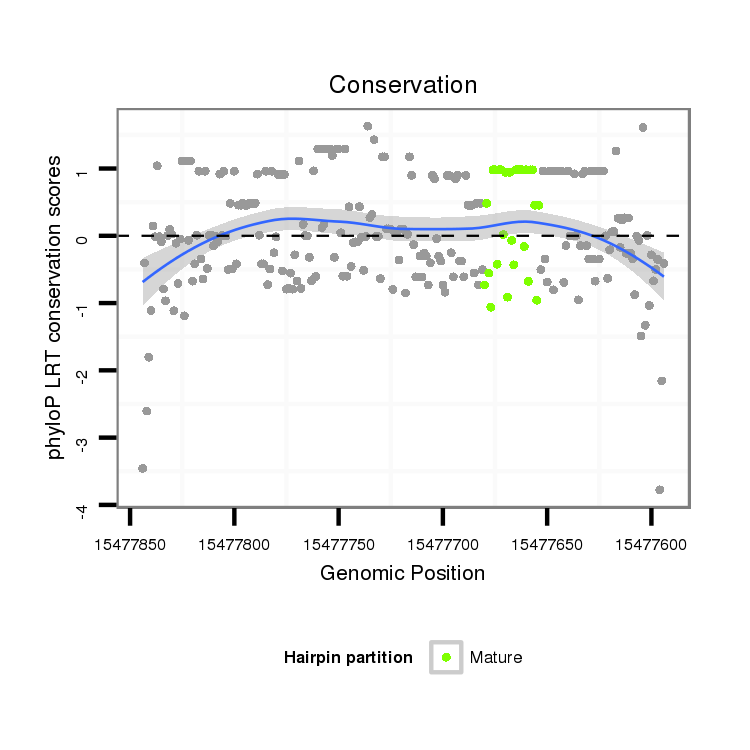
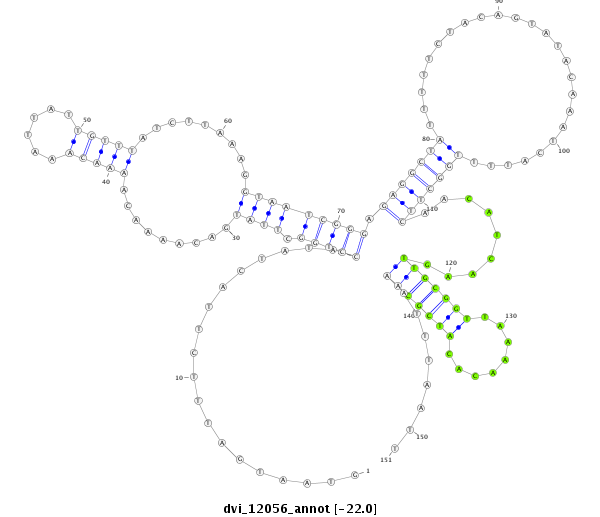
ID:dvi_12056 |
Coordinate:scaffold_12963:15477644-15477794 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.2 | -22.0 | -21.9 |
 |
 |
 |
CDS [Dvir\GJ20893-cds]; exon [dvir_GLEANR_630:1]; intron [Dvir\GJ20893-in]
No Repeatable elements found
| --------##########################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GATTGAACATGGATGACAATTTTAGTTTAAACGACATCGATCTAAAGGAGGTAATGATTTCTTACTATACCTGGCTTATGACAAAACAAAACAAATTATTGTTTATCTTAAAGGTAATCGGGAGAGGCTATTTTTCTACAGTATACAAATCATTTTGGCTTCAACATCAAGTTGCGGTTAAAACACATCGCAAATTTAATTACGAATGGGCCAGAAATAAATACGCACAGCTATTAAGCATTGACCATAAG **************************************************...................(((((.((((.........((((((....)))))).........))))))))).(((((((.........................))))))).........(((((((........)))))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|
| ....GAACATGGCTGGCAATTTTAGT................................................................................................................................................................................................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................CATCAAGTTGCGGTTAAAACACATCGC............................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................TATTAAGCATTGACCATAA. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................ATCAAGTTGCGGTTAAAACACATCGCAA.......................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................ATATAAAGGAGATAATGAT................................................................................................................................................................................................. | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................AAACGCTATTTTTCTACCGT............................................................................................................. | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
CTAACTTGTACCTACTGTTAAAATCAAATTTGCTGTAGCTAGATTTCCTCCATTACTAAAGAATGATATGGACCGAATACTGTTTTGTTTTGTTTAATAACAAATAGAATTTCCATTAGCCCTCTCCGATAAAAAGATGTCATATGTTTAGTAAAACCGAAGTTGTAGTTCAACGCCAATTTTGTGTAGCGTTTAAATTAATGCTTACCCGGTCTTTATTTATGCGTGTCGATAATTCGTAACTGGTATTC
**************************************************...................(((((.((((.........((((((....)))))).........))))))))).(((((((.........................))))))).........(((((((........)))))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060656 9x160_ovaries_total |
SRR060660 Argentina_ovaries_total |
SRR060678 9x140_testes_total |
SRR060689 160x9_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060665 9_females_carcasses_total |
SRR060670 9_testes_total |
SRR060681 Argx9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................ATGCTATGGACAGAATACT.......................................................................................................................................................................... | 19 | 2 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TGCTATGGACAGAATACT.......................................................................................................................................................................... | 18 | 2 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AATGCTATGGACAGAATACT.......................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TGCTATGGACAGAATACTG......................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TGTCATAGGTTTAGTAAT................................................................................................ | 18 | 2 | 12 | 0.75 | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TGTCATAGGTTTAGTAATGC.............................................................................................. | 20 | 3 | 16 | 0.56 | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GTATGCTATGGACGGAATACT.......................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................CTGTCATAGGTTTAGTAA................................................................................................. | 18 | 2 | 10 | 0.30 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AATGCTATGGACAGAATA............................................................................................................................................................................ | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................TATGCTATGGACGGAATACT.......................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................GAAGATCTAGTTCAACGCAAA........................................................................ | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................CTGTCATAGGTTTAGTAAA................................................................................................ | 19 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GTATGCTATGGACCGAATAT........................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................ATGCTATGGACCGAATATG.......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:15477594-15477844 - | dvi_12056 | GATT------GAACATGGATGACAATTTTA--GTTTAAACGACATCGATCTAAAGGAGGTAATGATTTCTTACTATACCTGGCTTATGACAAAACAAAACAAATTAT----------TGTTTATCTTAAAGGTAATCGGGAGAGGCTATTTTTCTACAGTATACAAATCATTTTGGCTTCAACATCAAGTTGCGGTTAAAACACATCGCAA------ATTTAATTACGAATGGGCCAGAAATAAATACGCACAGCTATTAAGCATTGACCATAAG |
| droMoj3 | scaffold_6500:18433195-18433446 + | AATT------TAACATGTGTGCGGATGTGA--AATTGCAAGATATCGATTTTATGGAGGTAATGAAATAATA----------TTTTA-----AAATAAACGAATAAATTGTGAGAGTTTGCTACTTTAAAGGAAATCGTGAAAGATTATGGCGCCGTAGTGAGAAAGGCAAAGTGGCATAAGAAAGAAATTGCCGTAAAAATATATCCCGATATTCTGATTGATTATAAATGGTTCAAGAGCAGATATGAGCAGCTATTGAATGTCGATCATGAG | |
| droGri2 | scaffold_14624:2201195-2201232 + | AAAAAGAGAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACACGACAGTCATGCTTTGACCATTCG | |
| droWil2 | scf2_1100000004521:2714845-2714926 - | AAAACAGGAT-----------------------------------------------------------------------------------------------------------TATTAAACCTAAAAACAACTGGGCATGCTTAGTTTTGTTTAGAATTTAAGTCATTTT--------------------------------------------------------------------------------------GGCCTACCCAACAAG | |
| dp5 | 2:5936145-5936172 - | CACA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCTGTCGCGAGTGGCCCATGAG | |
| droPer2 | scaffold_2:319578-319624 - | AAAAAATTAT-----------------------------------------------------------------------------------------------------------TATTTATATTAAAAATAATCATTAGAAGTT-------------------------------------------------------------------------------------------------------------------------CTTTATC | |
| droKik1 | scf7180000302299:6892-6941 + | ACAACTGGTT--------------------------------------------------------------------------CA-AAAACAACAAAACGAATTAT----------TGCCGATTTTCGA--------------------------------------------------------------------------------------------------------------------------------------------GGTAA | |
| droEle1 | scf7180000490995:93274-93303 + | GGTT------AAACATGGACTACATTTTTA--G---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCCAA | |
| droRho1 | scf7180000777274:4167-4212 + | TTTC------TT-----------------------------------------------------------CTTATAATATGCTGATATTAAAACAAAACAAAATAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTT | |
| droBia1 | scf7180000302127:67817-67895 - | TAGGATACAT------C-----------------------------------------------------------------------------------------------------GTTTCTTAT-----------------------------------------------------------------------------------A------ACTTCATTAAGATAGACCCAGAAAAAAATTGACACAATTACTAAACATCCGCTTCCAA | |
| droTak1 | scf7180000412945:559-658 - | ATTT------TAGCTGAAACTTCGATGTTATAATCTAAACAGCTGCAAA--TAA---------GGTTTAATATTAAATATAATTTAT-----AACAAAACAAAAAAACTTTA-----G----------------------------------------------------------------------------------------------------------------------------------------------------GTTTATATT | |
| droYak3 | 3R:20377978-20378014 - | CAACAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAATTACCGGTAAAACAAAT-----------------------------------------------------------TGAAAATTTA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/19/2015 at 05:07 PM