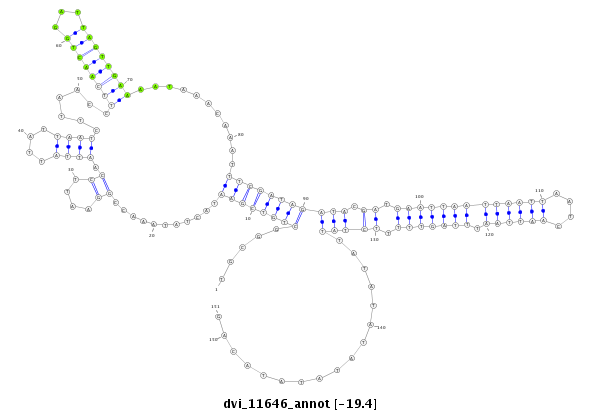
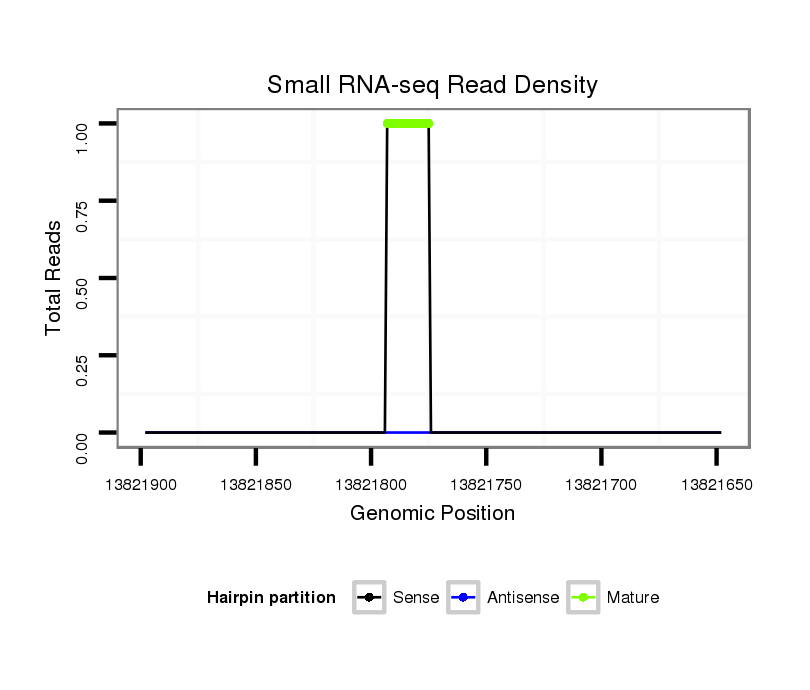
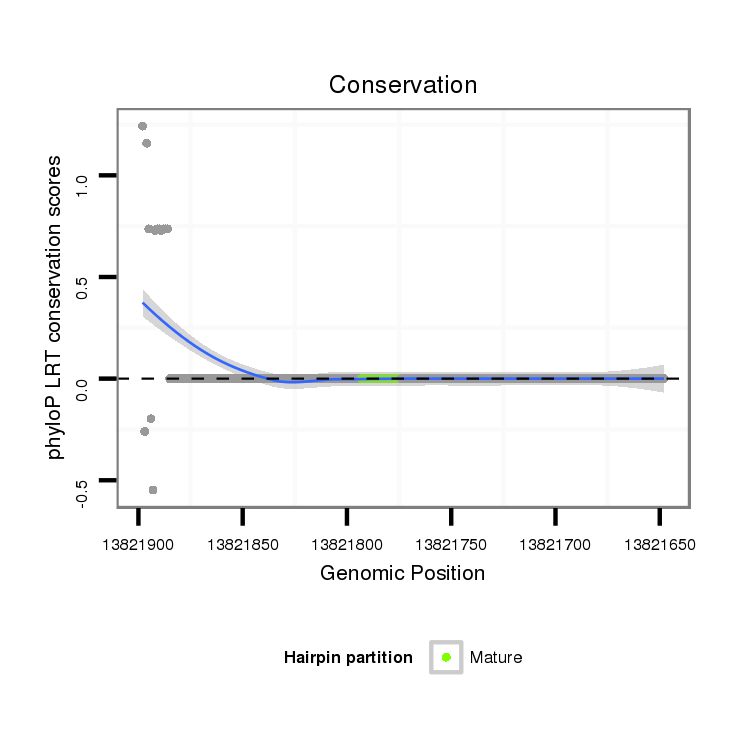
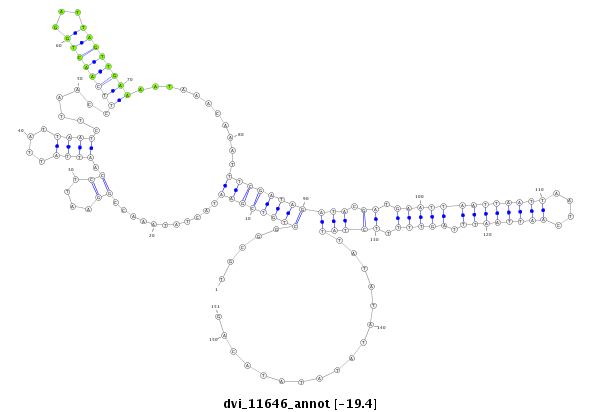
ID:dvi_11646 |
Coordinate:scaffold_12963:13821698-13821848 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
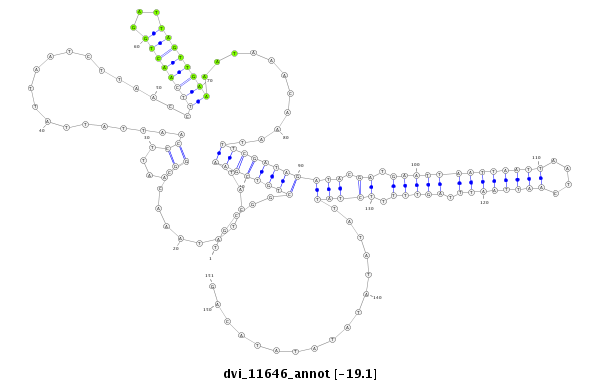
| -19.4 | -19.4 | -19.1 |
 |
 |
 |
CDS [Dvir\GJ22237-cds]; exon [dvir_GLEANR_754:3]; intron [Dvir\GJ22237-in]
| Name | Class | Family | Strand |
| (TTAA)n | Simple_repeat | Simple_repeat | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------###########################----------------------- TGCAAGCAAGAAAAAATCCTGATACATTAAAAGTAAGTTCAGATGAATTTTGCGGCTGTCGAATACTATAAACCGGAATTCCAATTATTATTAATCTTAACCTTCAACTGGATTAGTTGAAAATAAACAAATTTCGATAGATACGATGAATTAATTAATTAATCAATTAATTTAGTTTTTCTATTATATATATATATACAGTTCTTTGATTCCAAGTCCTTGATCTAACATTTGGTTAACCTGCCTATTTT **************************************************.....((((((((...........((....)).((((....)))).......((((((((...))))))))...........))))))))(((.((.(((((((((((((....)))))).))))))).))))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060669 160x9_females_carcasses_total |
SRR060657 140_testes_total |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
M027 male body |
SRR060687 9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................TTTTGCGGCTGTCGGAT........................................................................................................................................................................................... | 17 | 1 | 3 | 1.33 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AACTGGATTAGTTGAAAAT............................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATACTATACACCGGAATTTC......................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GAATACTATAAACCGGAATGC.......................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................GAATATAAAGAAATTTCGAT................................................................................................................. | 20 | 2 | 14 | 1.00 | 14 | 0 | 0 | 0 | 0 | 3 | 10 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................TAACACGTGGGTAACCTGCCT..... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TTTTGCGGCTGTCGGATC.......................................................................................................................................................................................... | 18 | 2 | 9 | 0.33 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................AACACGTGGGTAACCTGCC...... | 19 | 3 | 18 | 0.28 | 5 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................TGGTTTTTGCGGCTGTCGGAT........................................................................................................................................................................................... | 21 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TAACACGTGGGTAACCTGC....... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................TGATGGTTTTTGCGGCTGTC............................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................TTTTGCGGCTGTCAGAT........................................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................GAATTTTGCGGCTGTCTG............................................................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................CAGGAAGTTCAGCTGAATT.......................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ACGTTCGTTCTTTTTTAGGACTATGTAATTTTCATTCAAGTCTACTTAAAACGCCGACAGCTTATGATATTTGGCCTTAAGGTTAATAATAATTAGAATTGGAAGTTGACCTAATCAACTTTTATTTGTTTAAAGCTATCTATGCTACTTAATTAATTAATTAGTTAATTAAATCAAAAAGATAATATATATATATATGTCAAGAAACTAAGGTTCAGGAACTAGATTGTAAACCAATTGGACGGATAAAA
**************************************************.....((((((((...........((....)).((((....)))).......((((((((...))))))))...........))))))))(((.((.(((((((((((((....)))))).))))))).))))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
V116 male body |
GSM1528803 follicle cells |
M047 female body |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................AGAATTGGAACGTGCCCTA.......................................................................................................................................... | 19 | 3 | 18 | 5.00 | 90 | 27 | 27 | 11 | 9 | 1 | 3 | 0 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 |
| ..............................................................................................AGAATTGGAACGTGCCCT........................................................................................................................................... | 18 | 3 | 20 | 0.85 | 17 | 11 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GTTCTTTTGTAGGTCTATGTA................................................................................................................................................................................................................................ | 21 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................TAATCTTCATTCAAG................................................................................................................................................................................................................... | 15 | 1 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................TAAGGCTAATAATAGATAGA.......................................................................................................................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................AAAATTAGAATTGGAACGTG............................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................AGAATTGGAATGTGCCCTA.......................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:13821648-13821898 - | dvi_11646 | TGCAAGCAAGAAAAAATCCTGATACATTAAAAGTAAGTTCAGATGAATTTTGCGGCTGTCGAATACTATAAACCGGAATTCCAATTATTATTAATCTTAACCTTCAACTGGATTAGTTGAAAATAAACAAATTTCGATAGATACGATGAATTAATTAATTAATCAATTAATTTAGTTTTTCTATTATATATATATATACAGTTCTTTGATTCCAAGTCCTTGATCTAACATTTGGTTAACCTGCCTATTTT |
| dp5 | 4_group3:4034873-4034875 + | TAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396568:744101-744113 - | TGCAGCCAAGAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779970:18966-18968 - | TAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/19/2015 at 05:00 PM