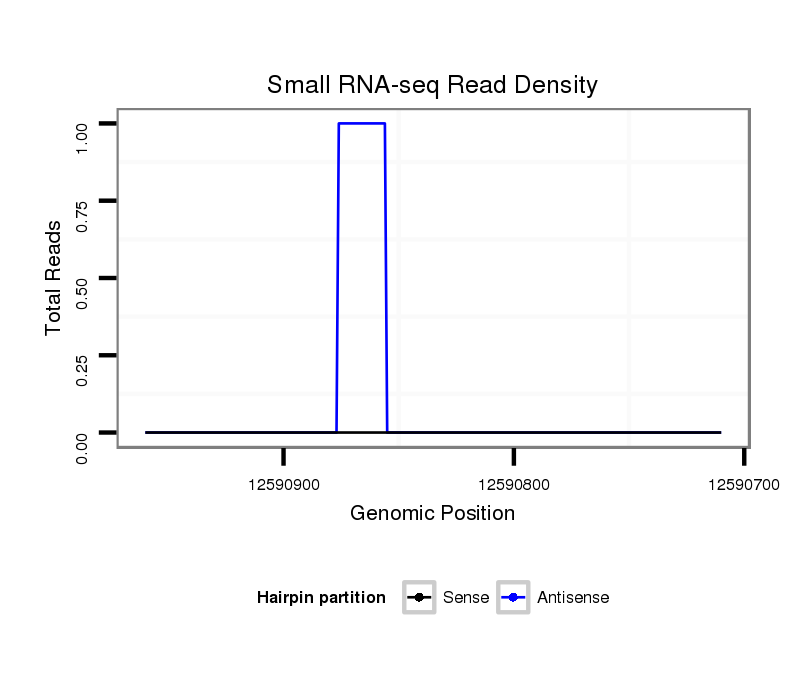
ID:dvi_11501 |
Coordinate:scaffold_12963:12590760-12590910 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ22919-cds]; exon [dvir_GLEANR_828:1]; intron [Dvir\GJ22919-in]
| Name | Class | Family | Strand |
| Helitron-1N1_DVir | RC | Helitron | - |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAAGATATTGCCTTGCGTTAGTCTCAAATTGTGTTTAAGGCATGCGATTGGTGAGTTTACAAAACAGGATTTTAAAAATTTTTATAAAACATTCATCTCCTTTAAGAGCTTTAAACTTGACATTTGAAAGGTAGAAGCTTCTATTGAAACCTCAGACGACGCCTTTTTCAAATACTTGATAACTTATTCACCATATTTGAGAGAGTATAAAAGTACTCTTTGTATTCTGTGCGAATTTTAAAATCTTTAAA **************************************************.(((((((.(((...((((............................))))....(((((((...(((.........)))...)))))))...)))))).))))............((((((((..(((........)))...))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060667 160_females_carcasses_total |
SRR060680 9xArg_testes_total |
V047 embryo |
SRR060659 Argentina_testes_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................GAAGCTTCTGTTGATACC.................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................AAGGTGGAAGCTTCTACT.......................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AGAAGCTTCTGTTGACACCA................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................GGGCCATAAACTTGACATTTG............................................................................................................................. | 21 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................AAGAGAGTAAAAAAGTAC................................... | 18 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................TGGTTAAGGCCTGCGACTG......................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TTTCTATAACGGAACGCAATCAGAGTTTAACACAAATTCCGTACGCTAACCACTCAAATGTTTTGTCCTAAAATTTTTAAAAATATTTTGTAAGTAGAGGAAATTCTCGAAATTTGAACTGTAAACTTTCCATCTTCGAAGATAACTTTGGAGTCTGCTGCGGAAAAAGTTTATGAACTATTGAATAAGTGGTATAAACTCTCTCATATTTTCATGAGAAACATAAGACACGCTTAAAATTTTAGAAATTT
**************************************************.(((((((.(((...((((............................))))....(((((((...(((.........)))...)))))))...)))))).))))............((((((((..(((........)))...))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060672 9x160_females_carcasses_total |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|
| ....................................................................................ATTTTGTAAGTAGAGGAAATT.................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| ......................................................................................................................................................................................................................................CGCTTAAAATCTTAGAAA... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:12590710-12590960 - | dvi_11501 | AAAGATATTGCCTTGCGTTAGTCTCAAATTGTGTTTAAGGCATGCGATTGGTGAGTTTACAAAA----CAGGATTTTA---AAAATTTTTATAAAACATTCATCTCCTTTAAGAGCTTTAAACTTGACATTTGAAAGGTAGAAGCTTCTATTGAAACCTCAGACGACGCCTTTTTCAAATACTTGATAACTTATTCACCATATTTGAGAGAGTATAAAAGTACTCTTTGTATTCTGTGCGAATTTTAAAATCTTTAAA |
| droMoj3 | scaffold_6500:29341467-29341566 + | AAAGAGTTTGCCCCGCGGCATACTTAAATTGTGTTTGACGTATGAGATTGGTGAGTCCACAAAACCTACAGCATCTTA---GCAACTCGCATTAACAGTGCAT----------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15252:175523-175609 + | AAAGATCTTGCCACGAGTCAGTTTCAAAGTGGGTTTTAATCATGCGAATGGTAAGTCTATAAAT----TAG---TTTA---GCAACTTTTAAGA------CAC----------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415779:506917-507001 + | AAGA-GCTTTCCCTCTGTCGAAGTAAAGTTGTGTTTCGGTGAAGCAAAGGGTAAGAATACTATT----TGA---TTTTCCCAAAAGTTCTATA--------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/17/2015 at 04:47 AM