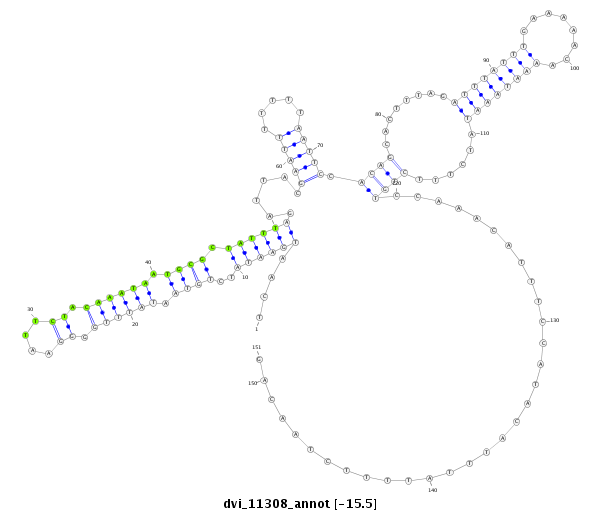
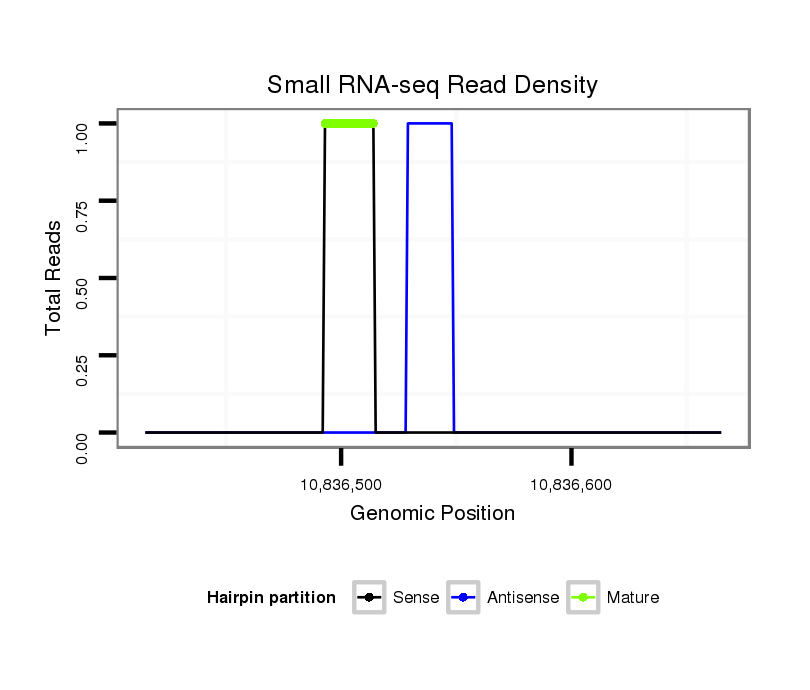
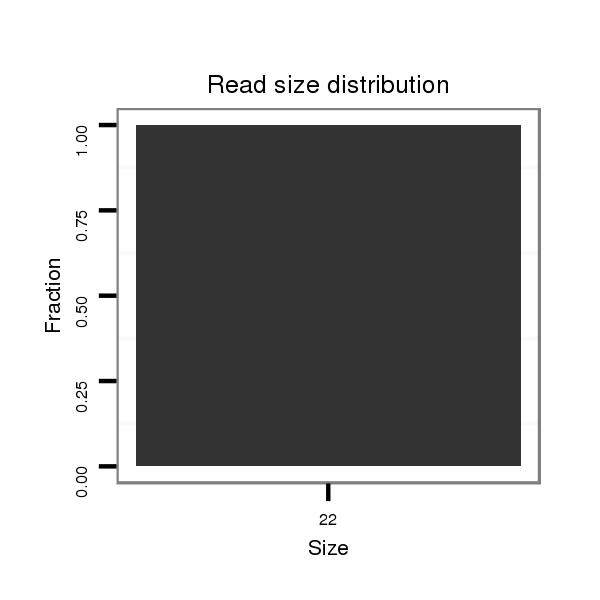
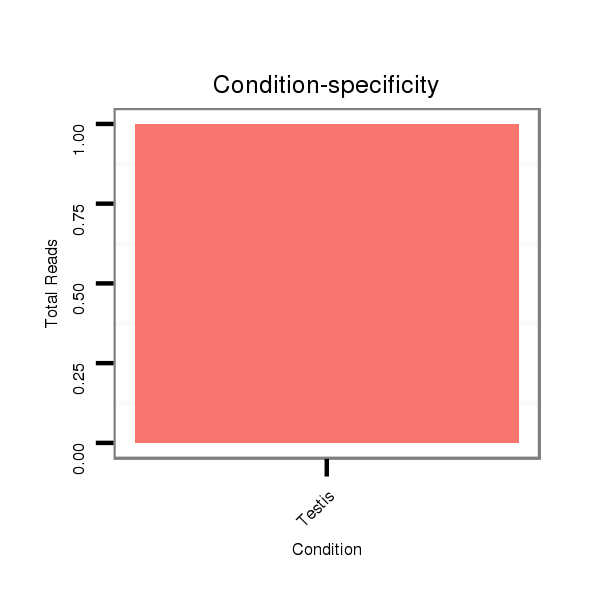
ID:dvi_11308 |
Coordinate:scaffold_12963:10836465-10836615 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -14.7 | -14.6 | -14.6 |
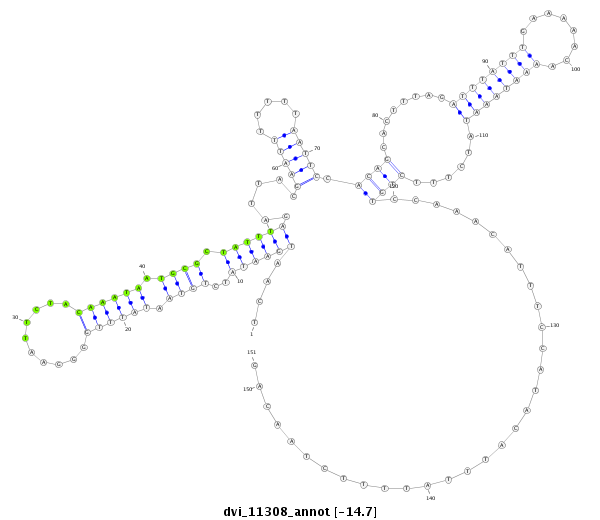 |
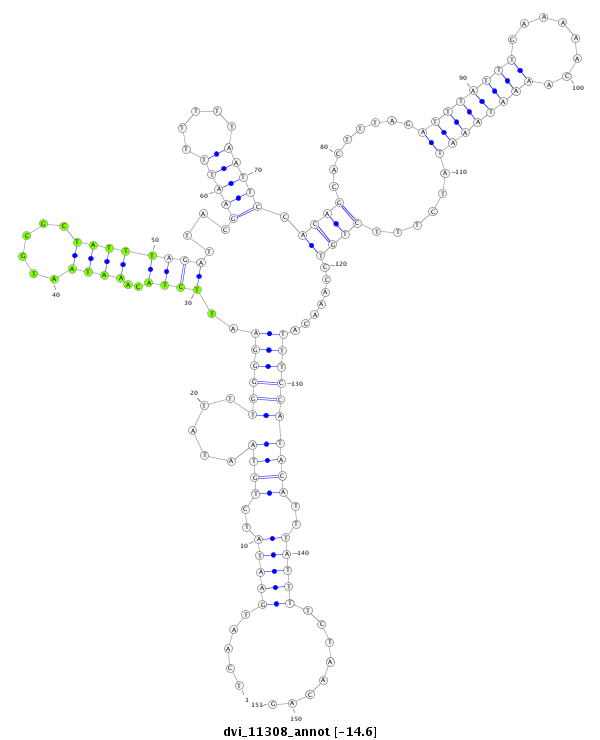 |
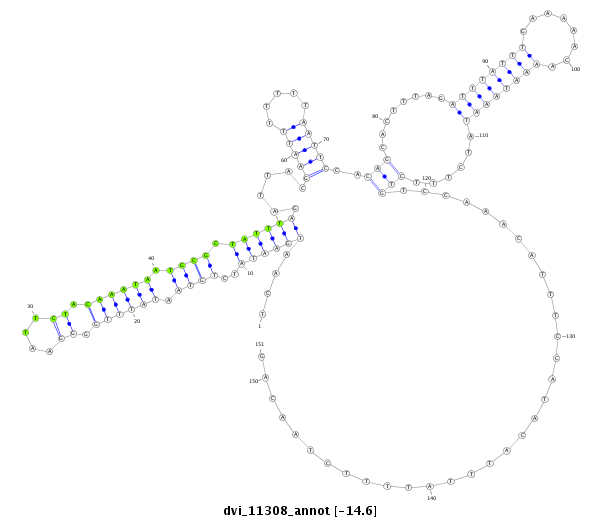 |
CDS [Dvir\GJ17346-cds]; exon [dvir_GLEANR_1947:4]; intron [Dvir\GJ17346-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACAGAATTGAATACTCCATAATATAAAGCAACTGACCCAATAAAATTCCATCAATGAATATCTGTAATATTTGGGGAATTCTACAAATAATGCGCTATTTAGATTACGAATTTTTTTAATTCCACAGCACTTTAGATTTATTTGAAAAACAAAATAAATATCTTTCTGTCCAAACATTTCCATACATTTATTTTCTAACAGGAAATCTTAGATCAGGTGGAATATTTTAAGGCAGACAATCTCGCGGAAGT **************************************************....((((((..((((.((((((.((....)).)))))).)))).))))))......(((((.....))))).((((........((((((((........))))))))......))))................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
V116 male body |
|---|---|---|---|---|---|---|---|
| ..............................................................................TTCTACAAATAATGCGCTATTT....................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................TCTGTAATGTTTGGGGGAT............................................................................................................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 |
|
TGTCTTAACTTATGAGGTATTATATTTCGTTGACTGGGTTATTTTAAGGTAGTTACTTATAGACATTATAAACCCCTTAAGATGTTTATTACGCGATAAATCTAATGCTTAAAAAAATTAAGGTGTCGTGAAATCTAAATAAACTTTTTGTTTTATTTATAGAAAGACAGGTTTGTAAAGGTATGTAAATAAAAGATTGTCCTTTAGAATCTAGTCCACCTTATAAAATTCCGTCTGTTAGAGCGCCTTCA
**************************************************....((((((..((((.((((((.((....)).)))))).)))).))))))......(((((.....))))).((((........((((((((........))))))))......))))................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060672 9x160_females_carcasses_total |
SRR060665 9_females_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060664 9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................AAATTAAGGTGTCGTGAAAT..................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTTTTGCTTTATTTAGAGAAA...................................................................................... | 21 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................TTTCTTTGACTGGGTTACT................................................................................................................................................................................................................ | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................GTTATCTTAAAGTAGTTAC................................................................................................................................................................................................... | 19 | 2 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ..........TGTGAGGTATTATACTTC............................................................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTTTTGCTTTATTTAGAGAAAC..................................................................................... | 22 | 3 | 13 | 0.15 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................TTTTGCTTTATTTAGAGAAACA.................................................................................... | 22 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................TTTTTTGCTTTATTTAGAGAAA...................................................................................... | 22 | 3 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................TTTTGCTTTATTTAGAGAAA...................................................................................... | 20 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:10836415-10836665 + | dvi_11308 | ACAGAATTGAATACTCCATAATATAAAGCAACTGACCCAATAAAATTCCATCAATGAATATCTGTAATATTTGGGGAATTCTACAAATAATGCGCTATTTAGATTACGAATTTTTTTAATTCCACAGCACTTTAGATTTATTTGAAAAACAAAATAAATATCTTTCTGTCCAAACATTTCCATACATTTATTTTCTAACAGGAAATCTTAGATCAGGTGGAATATTTTAAGGCAGACAATCTCGCGGAAGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/16/2015 at 11:01 PM