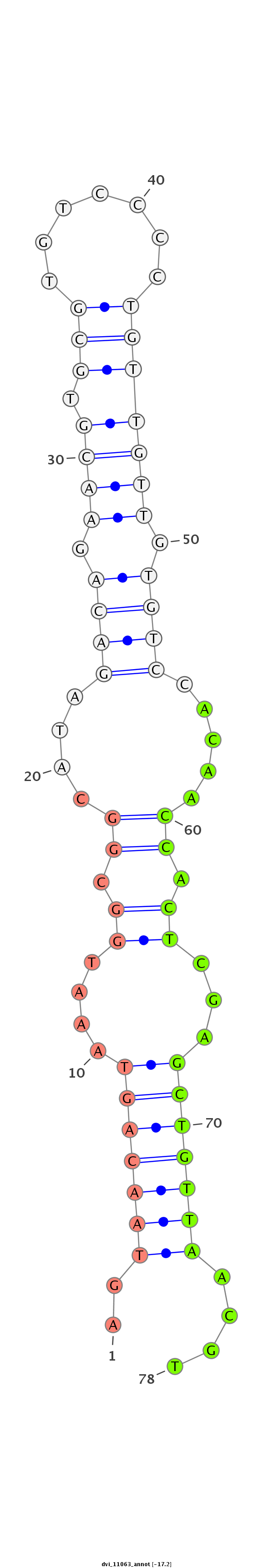
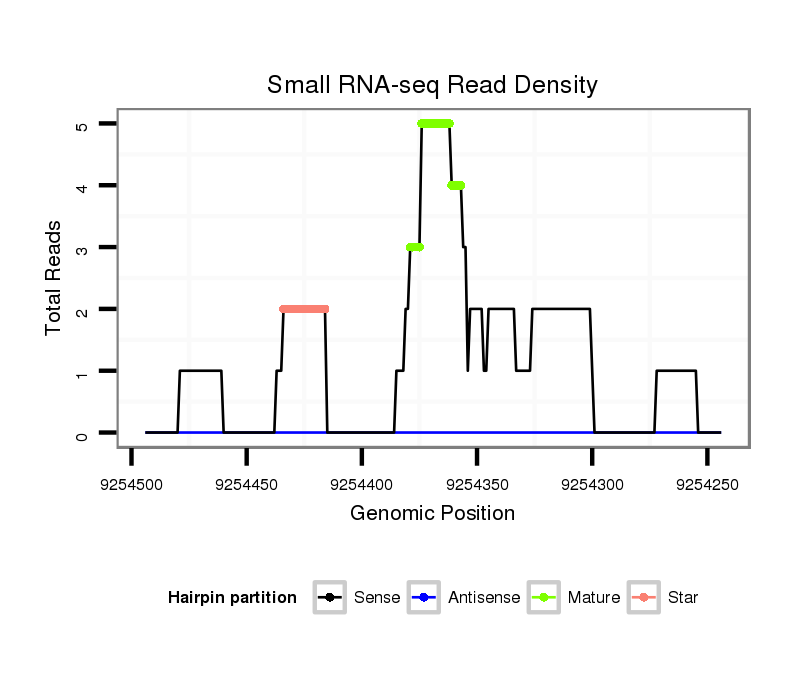
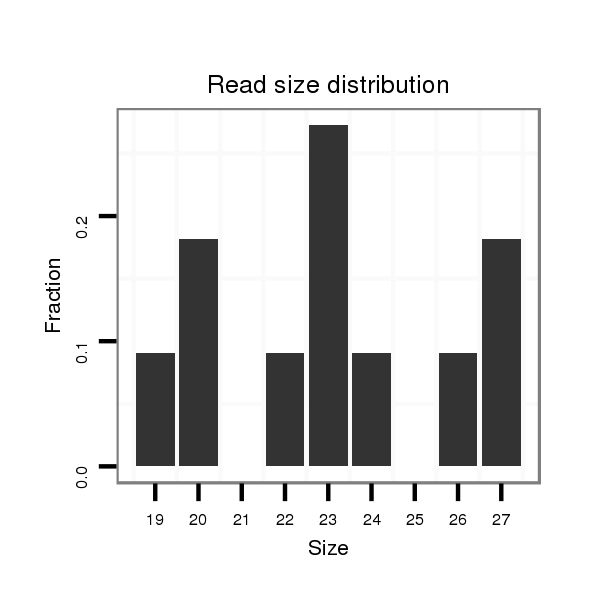
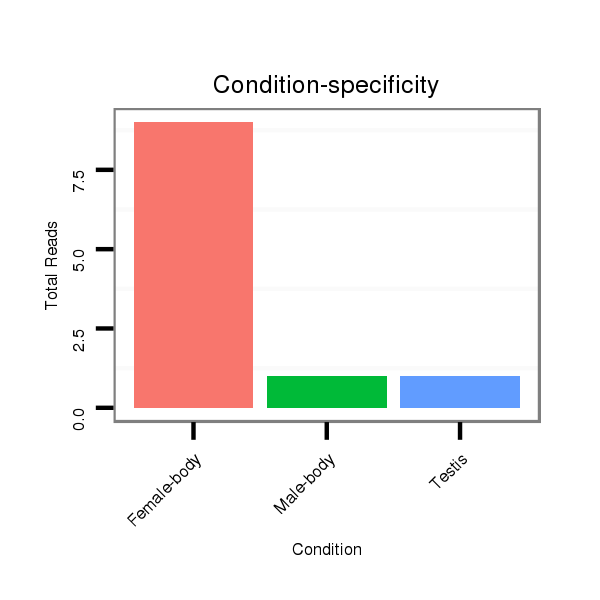
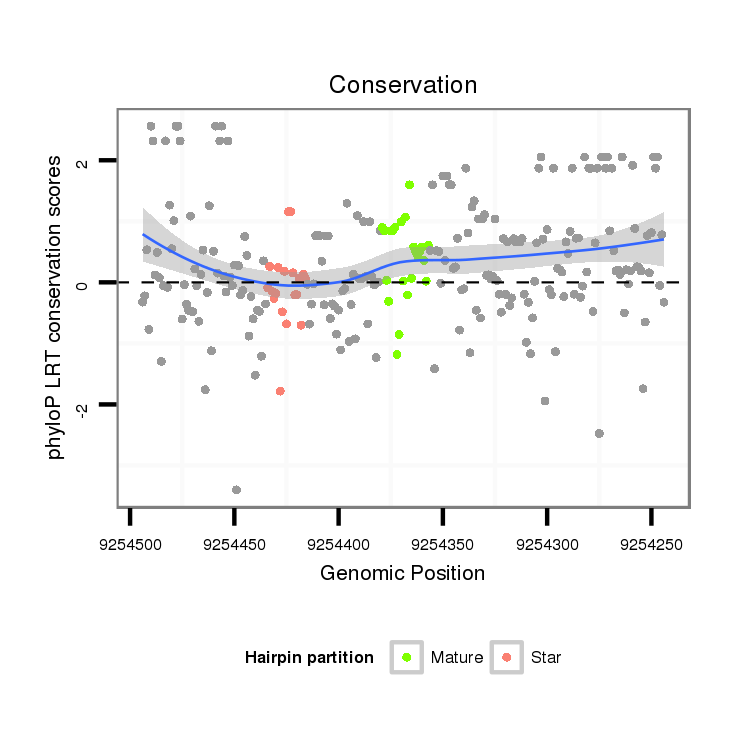
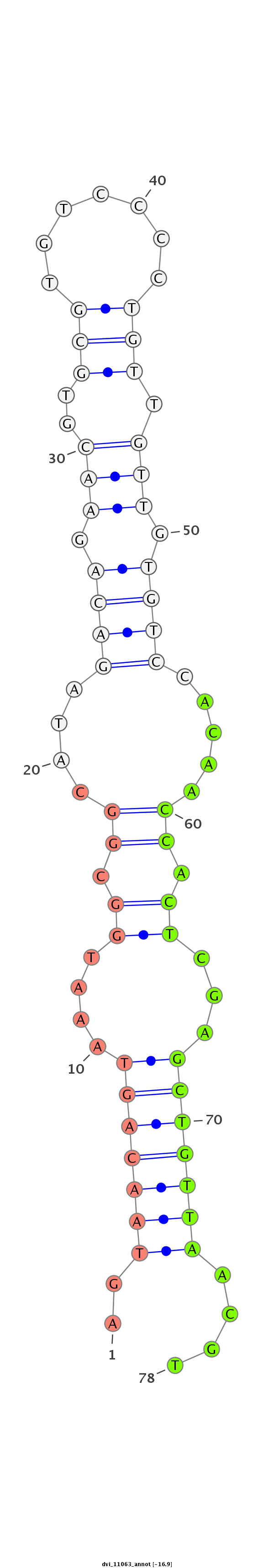
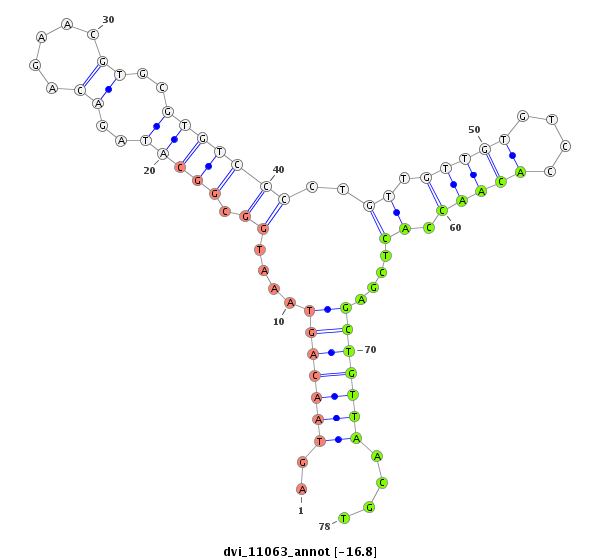
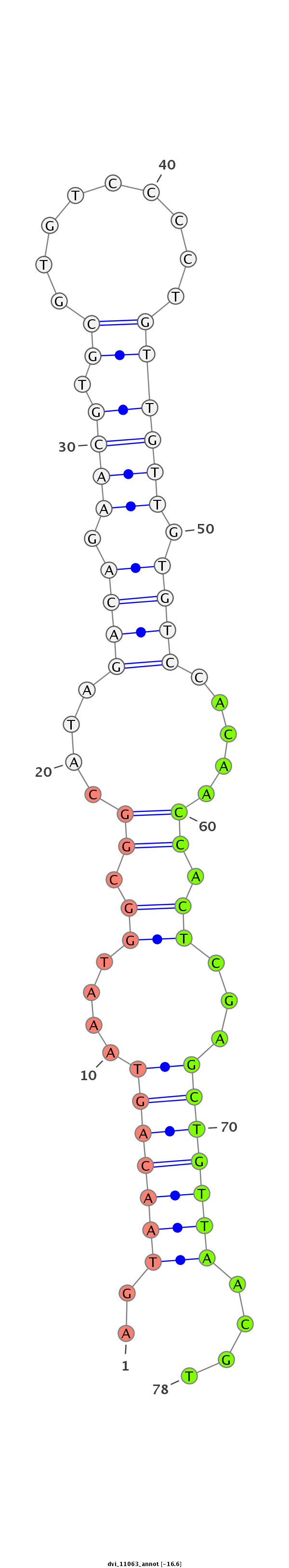
| CAGATGCCGTCGCATGATGTCCAGAAACAACATTGTGGTGACCGTGCCAGCAGGCCAGGTGGCTTGTCCAGTGCA----------------------G---ACCACAGCCA-----------------------------------------------CCACAGTGATGCCTGCCCAGTGTGCAACCCACTGGCAGTC--TCA------------------------------------------------------------------------------------------------------GCAGCAG---TCTCAGACAATGTCCGCACTGCCACCCAAGTACGA---TCAGGCTGTGGCAGCGAACTGAG-- | Size | Hit Count | Total Norm | Total | M055
Female-body | M057
Embryo | V060
Head | V040
Embryo | V107
Male-body | V108
Head | GSM1528801
follicle cells |
| ..................................................................................................................................................................................................................................................................................................................................................ACCCAAGTACGA---TCAGGCTGTGGC............. | 24 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTCCAGTGCA----------------------G---ACCACAGC............................................................................................................................................................................................................................................................................. | 20 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .AGATGCCGTCGCATGATGTCC.................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................CACCCAAGTACGA---TCAGGCTGTGGC............. | 25 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................AGACAATGTCCGCACTGC......................................... | 18 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| .................................................................................................................................................................................GTGTGCAACCCACTGGCAGT..................................................................................................................................................................................... | 20 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .AGATGCCGTCGCATGATGTCCA................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................ACCCAAGTACGA---TCAGGCTGT................ | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GATGCCGTCGCATGATGTCC.................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................GTGTGCAACCCACTGGCAGTC--TCA------------------------------------------------------------------------------------------------------GC....................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................GTGCCAGCAGGCCAGGTGGCTT......................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CA------------------------------------------------------------------------------------------------------GCAGCAG---TCTCAGACAATGTCCGCA............................................. | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AGAAACAACATTGTGGTGACCGTGCCA......................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CAGAAACAACATTGTGGTGACC............................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GTCCAGTGCA----------------------G---ACCACAGCCA-----------------------------------------------CCACA....................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................TCCAGAAACAACATTGTG..................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................GA---TCAGGCTGTGGCAGCGAACT..... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................C--TCA------------------------------------------------------------------------------------------------------GCAGCAG---TCTCAGACAAT.................................................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................GTGCAACCCACTGGCAGTC--T................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................AAGTACGA---TCAGGCTGTGGCAG........... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................GTACGA---TCAGGCTGTGGCA............ | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCACCCAAGTACGA---TCAGGCTGTG............... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................A------------------------------------------------------------------------------------------------------GCAGCAG---TCTCAGACAATGTCC................................................ | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................AATGTCCGCACTGCCACCCA................................... | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AGAAACAACATTGTGGTGACC............................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................GTACGA---TCAGGCTGTGGC............. | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................GTACGA---TCAGGCTGTGGCAGCGAA....... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TGCCAGCAGGCCAGGTGGCTTGTC...................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AGTC--TCA------------------------------------------------------------------------------------------------------GCAGCAG---TCTCA.......................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................TCAGGCTGTGGCAGCGAAC...... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................GA---TCAGGCTGTGGCAGCGAACTGA... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...ATGCCGTCGCATGATGTCCAGA................................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....TGCCGTCGCATGATGTCCAGA................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................GTCCAGAAACAACATTGT...................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................AGTACGA---TCAGGCTGTGGCA............ | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTCCAGTGCA----------------------G---ACCACAGCCA-----------------------------------------------CCACA....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................ACCCAAGTACGA---TCAGGCTGTGGCA............ | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........GTCGCATGATGTCCAGAAACAACATTGTG..................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................GCA----------------------G---ACCACAGCCA-----------------------------------------------CCACA....................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................GTGATGCCTGCCCAGTGTGCA.................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................GCAGCAG---TCTCAGACAATGTCCG............................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................ACCCAAGTACGA---TCAGGCTGTGGCAGC.......... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................GTACGA---TCAGGCTGTGGCAGC.......... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................CATTGTGGTGACCGTGCCAGCA...................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GTCCAGTGCA----------------------G---ACCACAGC............................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........GCATGATGTCCAGAAACAAC........................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................GA---TCAGGCTGTGGCAGCGA........ | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............TGATGTCCAGAAACAACATTGT...................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AGTC--TCA------------------------------------------------------------------------------------------------------GCAGCAG---TCTC........................................................... | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................ACAACATTGTGGTGACCGTGCCAGCA...................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................CACCCAAGTACGA---TCAGGCTGTGGCAGC.......... | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................G---ACCACAGCCA-----------------------------------------------CCACAGT..................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................CACCCAAGTACGA---TCAGGCTGTGGCA............ | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................GACCGTGCCAGCAGGCCAGGTG............................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................ATTGTGGTGACCGTGCCAGCA...................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................ACGA---TCAGGCTGTGGCAGCGAACTGA... | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................ACGA---TCAGGCTGTGGCAGCGAACTGAG-- | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................GTCCAGAAACAACATTGTGGTGACC............................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AAACAACATTGTGGTGAC................................................................................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................GGCAGTC--TCA------------------------------------------------------------------------------------------------------GCAGCAG---TCTCA.......................................................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................AGTGCA----------------------G---ACCACAGCCA-----------------------------------------------CCACAGT..................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................GCCACCCAAGTACGA---TCAGGCTGTG............... | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCACCCAAGTACGA---TCAGGCTGT................ | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................AGACAATGTCCGCACTGCCACCCA................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................TACGA---TCAGGCTGTGGCAGCGAACTGA... | 27 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................AAGTACGA---TCAGGCTGTGGCAGC.......... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CA------------------------------------------------------------------------------------------------------GCAGCAG---TCTCAGACAAT.................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................GCAACCCACTGGCAGTC--TCA------------------------------------------------------------------------------------------------------GC....................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................CCAAGTACGA---TCAGGCTGTGG.............. | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................ACATTGTGGTGACCGTGCCAGCA...................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................ACGA---TCAGGCTGTGGCAGCGAACT..... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................GTGATGCCTGCCCAGTGTGC................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................GTACGA---TCAGGCTGTGGCAGCGAAC...... | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................CATTGTGGTGACCGTGCCAG........................................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................GTACGA---TCAGGCTGTGGCAG........... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................CACAGTGATGCCTGCCCAGT....................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................GACAATGTCCGCACTGCC........................................ | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................CACCCAAGTACGA---TCAGGCT.................. | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................GA---TCAGGCTGTGGCAGCGAACTG.... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |