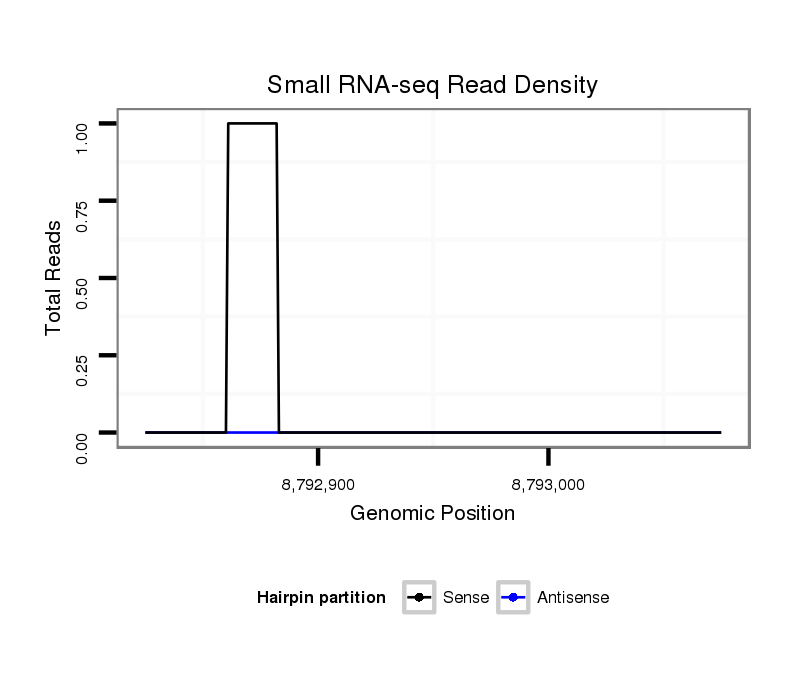
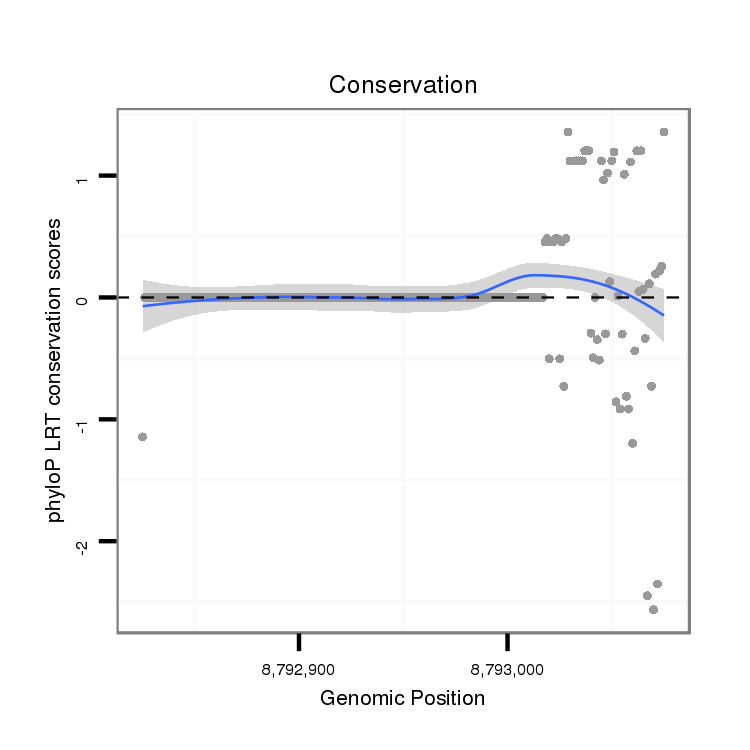
ID:dvi_11003 |
Coordinate:scaffold_12963:8792875-8793025 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ17241-cds]; exon [dvir_GLEANR_1845:2]; intron [Dvir\GJ17241-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GACTTACAAAAATGAAATATTTCTTTTAATATGTTTACCATAAAATTACAGTAGAGCCCAAACAATTAAAAAACCCATTATACCCGAAATTCTAGTTAATTTACCGAACTTGTTGAAATATTTGTACGAAACTGTGTGTCTGTGCTGAATGCCAGATTTAATGTGCAAGGCAGATAATTATGTGTCCGGGTGTGAGGCTAGGGAACGCCCCCAAAATACGGGGAACATCGCTTGGGCATATCGCAGGGCAA **************************************************...............(((......((.(((((((((......((((......................(((((((.....((..((.(((((.((.....))))))).))..)).....)))))))))))......))))))))))).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060667 160_females_carcasses_total |
SRR060669 160x9_females_carcasses_total |
M027 male body |
M028 head |
V047 embryo |
V053 head |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................GTATGAGGCCAGGGAACGC........................................... | 19 | 2 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................ATGAGGCCAGGGAACGCCCCT....................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GTATGAGGCCAGGGAACG............................................ | 18 | 2 | 3 | 1.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ....................................ACCATAAAATTACAGTAGAGCC................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................TGGAAGGTAGATAATTATGTCT.................................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................TGTATGAGGCCAGGGAACGC........................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................TGTCCAAGGTAGGTAATTAT...................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................ACGGGGAATATGGCTTCGG............... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................TGGTATGAGGCCAGGGAAC............................................. | 19 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................GATTATTATGTGTCGGGA............................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................GCAGCTAGAGAACGCCCCC....................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................GGCCAGGGAACGCCCCTCA..................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CTGAATGTTTTTACTTTATAAAGAAAATTATACAAATGGTATTTTAATGTCATCTCGGGTTTGTTAATTTTTTGGGTAATATGGGCTTTAAGATCAATTAAATGGCTTGAACAACTTTATAAACATGCTTTGACACACAGACACGACTTACGGTCTAAATTACACGTTCCGTCTATTAATACACAGGCCCACACTCCGATCCCTTGCGGGGGTTTTATGCCCCTTGTAGCGAACCCGTATAGCGTCCCGTT
**************************************************...............(((......((.(((((((((......((((......................(((((((.....((..((.(((((.((.....))))))).))..)).....)))))))))))......))))))))))).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060660 Argentina_ovaries_total |
SRR060681 Argx9_testes_total |
|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................TCCCTTGCCGGGGTTTCA.................................. | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 |
| .................................................................................................................................................................................................AGCGGATCCCTTGCGGGG........................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:8792825-8793075 + | dvi_11003 | GACTTACAAAAATGAAATATTTCTTTTAATATGTTTACCATAAAATTACAGTAGAGCCCAAACAATTAAAAAACCCATTATACCCGAAATTCTAGTTAATTTACCGAACTTGTTGAAATATTTGTACGAAACTGTGTGTCTGTGCTGAATGCCAGATTTAATGTGCAAGGCAGATAATTATGTGTCCGGGTGTGAGGCT-AGGGAACGCCCCCAAAATAC--GGGGAACATCGCTTGGGCATATCGCAGGGCAA |
| droMoj3 | scaffold_6500:2534129-2534184 - | T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAAGCTGAAGTAACGCCCCCAAAAC-TAATG---ACAGCTC--TGTCATAGCATCAGTCCA | |
| droGri2 | scaffold_15252:12993863-12993870 - | C------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A------------------------------------------TGGCAA | |
| dp5 | 4_group1:4296292-4296340 + | A------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACGCCCCCAAATA-CAGAGGGAACAAATTTCTGAAAAATCACAAACAAA | |
| droPer2 | scaffold_5:914269-914306 - | A------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACGCCCCCAAATA-CAGAGGGAAC-----------AAATTTCAGAACAA | |
| droFic1 | scf7180000453904:562714-562761 - | A------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACGCCCCCAAACA-TG-AGGCATCAGCAATGAGCGATATTCCTTGACAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
Generated: 05/17/2015 at 04:41 AM