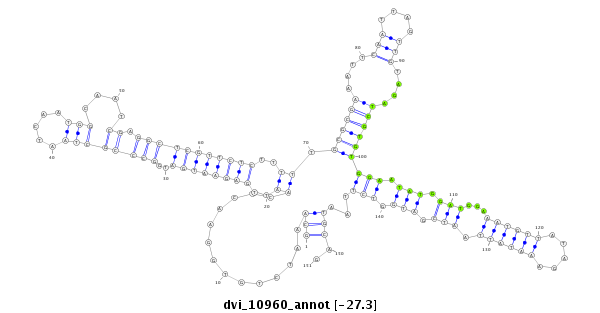
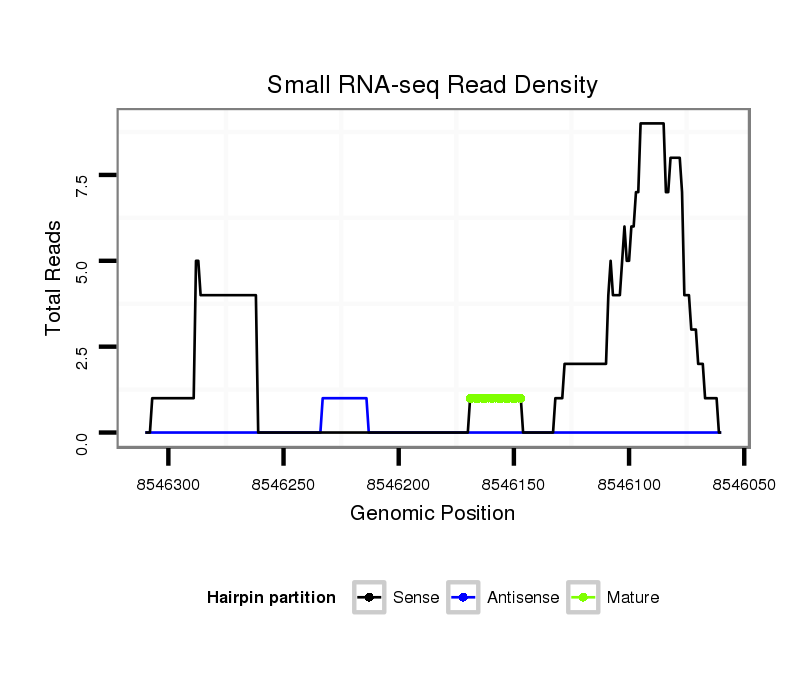
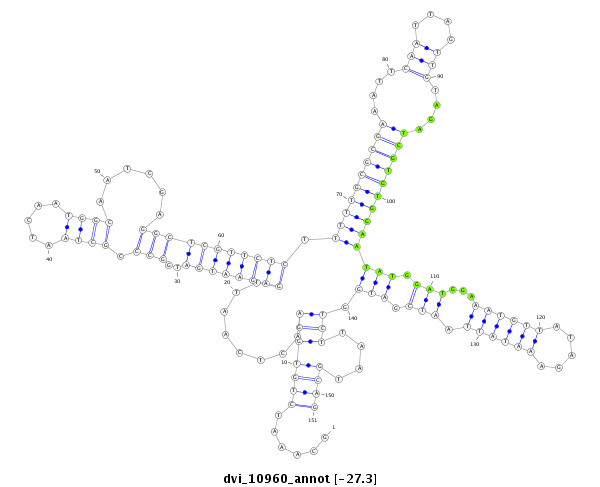
ID:dvi_10960 |
Coordinate:scaffold_12963:8546110-8546260 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
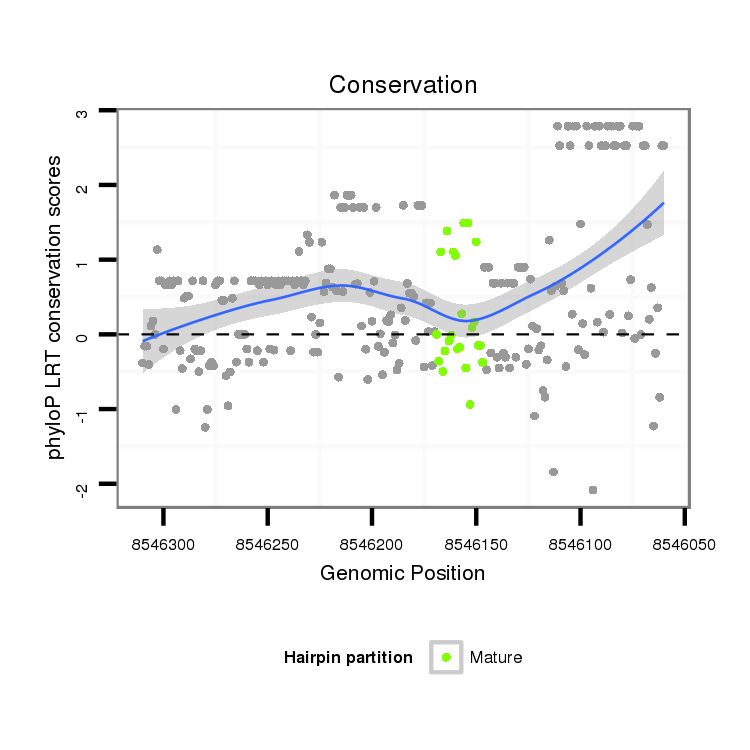
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.3 | -27.3 | -27.3 |
 |
 |
 |
exon [dvir_GLEANR_1011:2]; CDS [Dvir\GJ10166-cds]; intron [Dvir\GJ10166-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATTTGAAATTGGGACATTCGGTTCACATCAGTTGACATGCTGCTAGGTCTGCAAATCTGTGGAACTCAATTGAGAATGATGGCCCGCTAATCAATGGCAATCGAGGCTCGTTCTCTTTTTGCGCGAAATTCAATTAGTTGTAGATCGTGTGGAATATGGATGGAAATGTTATAGAAATATTAATCGATGGTCTTAATGCAGGAATGCAACACAACGCTGTGCGACTGCAAGGGCATCAAGGGACGCATGGG **************************************************(((..............((..((((((((..((((((((.....)))....)).)))))))))))..)).((((((....(((....)))....))))))(((.(((.(((...((((((.....)))))).))).))).)))...)))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
GSM1528803 follicle cells |
SRR060666 160_males_carcasses_total |
V047 embryo |
M027 male body |
SRR060667 160_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060683 160_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR1106713 embryo_0-2h |
V053 head |
SRR060689 160x9_testes_total |
SRR1106728 larvae |
SRR060682 9x140_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................TCACATCAGTTGACATGCTGCTAGGTC.......................................................................................................................................................................................................... | 27 | 0 | 1 | 4.00 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GAATGCAACACAACGCTGTGCGACT......................... | 25 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................ATTAATCGATGGTCTTAATGCAGGA................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................GCTGTGCGACTGCAAGGGC................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................ATCGATGGTCTTAATGCAGGAATGCAA.......................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...TGAAATTGGGACATTCGGTTC................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................ACACAACGCTGTGCGACTGCAAGGGC................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................GCTGTGCGACTGCAAGGGCATCAAG........... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GTAATGCAACACAACGCTGTGCGT........................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................CTGCAAGGGCATCAAGGGA........ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AATGCAACACAACGCTGTGCGA........................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................CAACGCTGTGCGACTGCAAGGGC................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AGATCGTGTGGAATATGGATGGA....................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................ACGCTGTGCGACTGCAAGGG.................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CAACACAACGATATGCGACTGCA...................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AAAGGAATGCAACACAACGCTGTGCGA........................... | 27 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................TTAGTTGTAGATCGTGTGGAATAAA............................................................................................. | 25 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................AAGGGCATCAAGGGACGCATG.. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................GCTGTGTGACTGCAAGGGCATC.............. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................AACACAACGCTGTGCGACTGCAAGGGCATC.............. | 30 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AAAGGAATGCAACACAACGCTGTGCGACT......................... | 29 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TGAGTATGACGGCACGCTA.................................................................................................................................................................. | 19 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................CATCAATTGAGATTCTGCTAG............................................................................................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........TGGGACATCCTGTTCACGTC.............................................................................................................................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................ACATCAATTGAGATTCTGCTA.............................................................................................................................................................................................................. | 21 | 3 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ......AATTGGGACCTTCGG...................................................................................................................................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............ATACGGTTCACATGAGTT.......................................................................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................GATGGTCTTATGGCTGGAA............................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TAAACTTTAACCCTGTAAGCCAAGTGTAGTCAACTGTACGACGATCCAGACGTTTAGACACCTTGAGTTAACTCTTACTACCGGGCGATTAGTTACCGTTAGCTCCGAGCAAGAGAAAAACGCGCTTTAAGTTAATCAACATCTAGCACACCTTATACCTACCTTTACAATATCTTTATAATTAGCTACCAGAATTACGTCCTTACGTTGTGTTGCGACACGCTGACGTTCCCGTAGTTCCCTGCGTACCC
**************************************************(((..............((..((((((((..((((((((.....)))....)).)))))))))))..)).((((((....(((....)))....))))))(((.(((.(((...((((((.....)))))).))).))).)))...)))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060660 Argentina_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
SRR060659 Argentina_testes_total |
SRR060684 140x9_0-2h_embryos_total |
M061 embryo |
SRR060675 140x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................CTACCGGGCGATTAGTTACC.......................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CGCGCTTAAAGTTAATCAAC............................................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................GCCGGGCGATTATTTAGCGTTA...................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TCGCGTTGCGAAACGCTGACGT...................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................CTGGCGATTATTTAGCGTTA...................................................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................TTAGTTACCGTTAGCCTC................................................................................................................................................. | 18 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................AGAGAAAAATGGGCTCTAA......................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................GGTGTAGATAACTGTACGA.................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:8546060-8546310 - | dvi_10960 | ATTTGAAATTGGGACATTCGGTTCACATC-------AGTTGACAT--------GCTGCTAGGTCTGCAAATCTGTGGAACTCAATTGAGAATGATGGCC-CGCTAATCAA---TGGCAATCGAGGCTCGTTCTCT-----TT--TT----------------GCGCGAAATTCAATTAGTTGTAGATCGTGTGGAATATGGAT--------------------------------------------------------------------------------------------------------------------GGAAATGTTATAGAAATATTAATC---------GA--TGGTC--TTAATGCAGGAATGCAACACAACGCTGTGCGACTGCAAGGGCATCAAGGGACGCATGGG-- |
| droMoj3 | scaffold_6500:1158336-1158646 - | ATTTGA-ATTGGGACAGTT----AACAGTTATAGGCATGAATCATATATCTATGCACTTAA----GTAAATCTATGGAACTCAATTAAGAATGATGAC--TGCTAATCAATCAAGGCAATCAAGGCGCGTTCTCC----------A----------------GTGCGAAATTCAATTACCCAT----T---------------------ATATGTATATATGTATATATACTTATGTCTTTTATTTGGATGTAGTTTCACAGATATATGAA-CGAAATCAA-----------------------------GTGAAT--CAATATT-------------TAATTTGTTTTTGT--T-TCTTTT-TTTTTTCAGGAATGCAATACAACGCTGTGCGACTGCAAAGGCATCAAAGGACGCATGGG-- | |
| droGri2 | scaffold_15252:11740695-11740961 - | ATTCGA-ATTAGGACAATCAGTTCATTTC-------ACAAATCAT--------------AA----ATAAAACTATGAAGCTCAATTGAGAATGATGACAGCGCTAATCAA---TGGCAATCAGGGCTCGATCGA--AG-------------------CG-----AAGAAATTCAATTAGTTGT----TG-----TATTTCGATTCAAATATTTGTAGATATATTTATA------------------------------ATAGTCATAAGAAGTGGAA---GCAGTTAGTCTC-ATGGCTTGTTA--------------------------------------------------------AC-CGATTTCAGACTTGCAACAACACGATATGCGACTGCAAGGGCATCAAGGGACGTATTGG-- | |
| droWil2 | scf2_1100000004585:8347232-8347351 - | T------------------------------------------------------------------------------------------------------TGGCG-----------------------------------------------------------------------------AATGCATGGAATAT-------------------------------------------------------------------------------CAA-----------------------------GTAAAT--CGGAATTATTTGAATACTAATAATT-------------TATATTTGATTTTTAGGAATGCAATAGTACAGTATGCGATTGCAAGGGCCTGAAGGGAAGAATTGG-- | |
| dp5 | 4_group4:1536912-1537067 - | C------------------------------------------------------------------------------------------------------TAATCAG---AGACAATCAAGACTTGTTCTCGAAT-------------------TAT-----TGAAACCCAATTCGG-GT----CG-----TCTGTCAATGCT-------------------------------------------------------------------------AACTGATGGACTCTACGATTCTCTAATTA-----------------------------------------------------T-CAATTTTAGGAATGCAATACAACGCTGTGTGACTGCAAGGGCATCAAAGGACGCATGGG-- | |
| droPer2 | scaffold_10:540803-540958 - | C------------------------------------------------------------------------------------------------------TAATCAG---AGACAATCAAGACTTGTTCTCGAAT-------------------TAT-----TGAAACCCAATTCGG-GT----CG-----TCTGTCAATGCT-------------------------------------------------------------------------AACTGATGGACTCTACGATTCTCTAATTA-----------------------------------------------------T-CAATATTAGGAATGCAATACAACGCTGTGTGACTGCAAGGGCATCAAAGGACGCATGGG-- | |
| droAna3 | scaffold_12916:13648374-13648432 - | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATTCCAGGAATGTAACACAACAGTATGTGACTGCAAAGGCATCAAAGGACGACAGGG-- | |
| droBip1 | scf7180000396535:618858-618912 - | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGAATGTAACACAACGGTATGTGACTGCAAAGGCATCAAAGGAAAACAGGGAG | |
| droKik1 | scf7180000302271:138585-138737 - | A-----------------------------------------------------------------------------------------A---TGATAGCTCCAATCAG---GGGCAATCAAGACCAGTTCTCGGACCGATTCCT----------------CTACGAAATTCGATTGGTTTC----------------------C--------------------------------------------------------------------AACTCAA-----------------------------CTGAAT--A-----------------------------------CTCCTTC--TTCTGGCAGGTCTGCAACACCACGCTATGTGACTGCAAGGGTATCAAGGGTCGCATCGG-- | |
| droFic1 | scf7180000453842:1231967-1232025 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATTCCAGATCTGCAACACAACGCTATGTGATTGCAAGGGAATTAAGGGACGAGTGGG-- | |
| droEle1 | scf7180000491273:216712-216764 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGATATGCAACACTACGCTATGCGACTGCAAGGGCATCAAAGGACGTGTTGG-- | |
| droRho1 | scf7180000777055:88391-88498 - | A--------------------------------------------------------------------------------------------------------ATCAG---GGGCAATCAAGACTAGTTCTAT-----T-----------------------TAAAAATTCGATTTGTTCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-TTGTTCCAGATCTGCAACGAAACGATATGCGACTGCAAGGGCCTTAAGGGATATGTTGG-- | |
| droBia1 | scf7180000302261:444744-444882 - | C------------------------------------------------------------------------------------------------------CAATCAG---GGGCAATCAAGACTCGCACA--------CTCCTTGG----------------C-ACATTTTATTAGTTCC--------------------TCG-------------------------------------------------------------------------AACTGAC------------------------GGGAAATGG-----------------------------------CACATTCT-ACTTTGCAGATCTGCAACACGACGGTGTGCGACTGCAAGGGCATCAAGGGACGCCAAGG-- | |
| droTak1 | scf7180000415204:133395-133570 + | ACCCAA-A--------------------------------------------------------------------------------------TGAGAGTTCCAATCAG---GGGCAATCAAGACCACTTCTAT-----ATTCCTTGGTATATATTCAGTGGTACAAAATTCTATTAGTTCC--------------------CCC-------------------------------------------------------------------------GACTGACTGGC-------------AAGTATC------------------------------------------AA--TTTTCT-TTATTCCAGATTTGCAATACGACGCTATGCGACTGCAAGGGCATCAAGGGACGCATGGG-- | |
| droEug1 | scf7180000409005:902561-902632 - | AAATGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-TATGCC-ACATTGCAGATATGTAACACAACGATATGTGACTGCAAGGGCATAAAGGGACGAGCCGG-- | |
| dm3 | chr2L:5018708-5018854 - | A--------------------------------------------------------------------------------------------------------ATCAG---AGGCAATCAAGCCTGGTTCCAT-----ATTCCTTGG-------------------GC--------------TACCGTGTGCTATATAGTT-TATACATATGTACATATATATATA--------------------------------------------------------------------------------------------------------------------------------TACGCT-TGATTTCAGATCTGCAACACAACGCTATGCGACTGCAAAGGAATCAAGGGACGCATGGG-- | |
| droSim2 | 2l:4831928-4831993 - | A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATTCT-TGATTTCAGATCTGCAACACAACGATATGCGACTGCAAAGGAATCAAGGGACGCATGGG-- | |
| droSec2 | scaffold_5:3105101-3105166 - | A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATTCT-TGATTTCAGATCTGCAACACAACGCTATGCGACTGCAAAGGAATCAAGGGACGCATGGG-- | |
| droYak3 | 2L:10507136-10507188 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGATCTGCAACACAACGCTATGCGACTGCAAGGGAATCAAGGGACGCATGGG-- | |
| droEre2 | scaffold_4929:5097154-5097206 - | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGATCTGTAATACAACGATATGCGATTGCAAAGGAATCAAGGGACGTATGGG-- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:18 AM