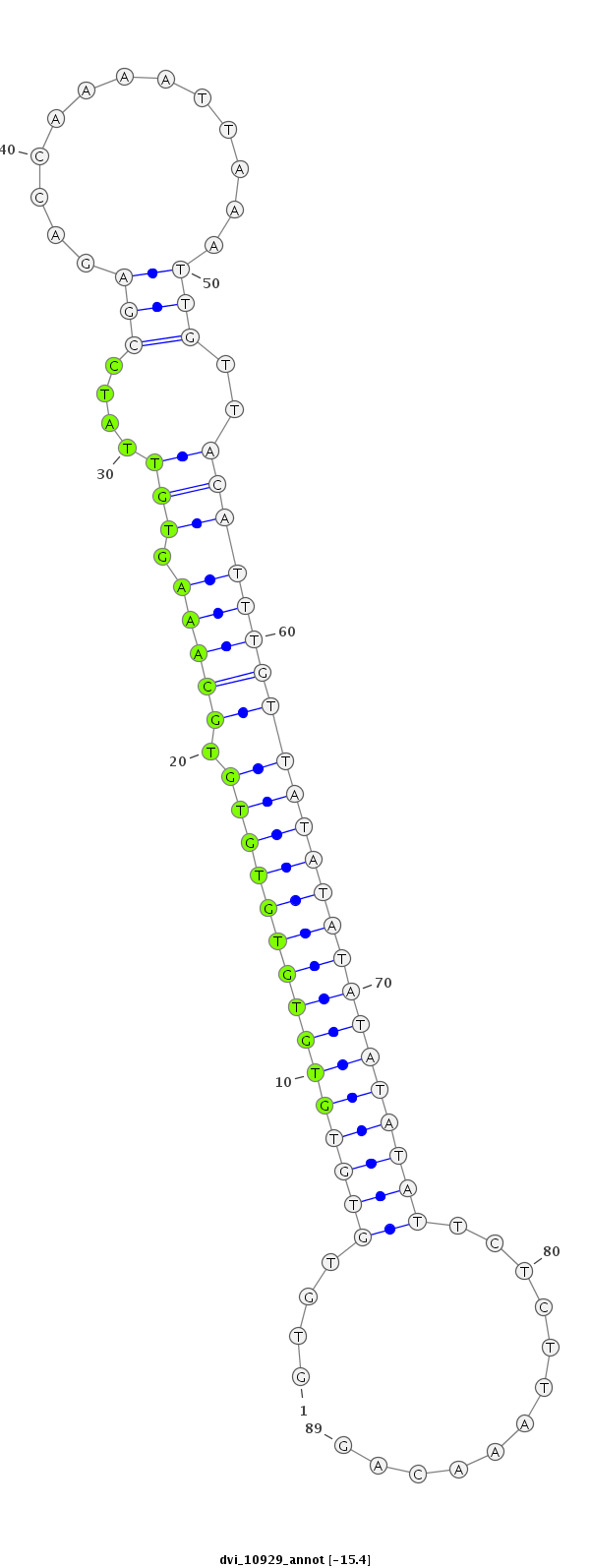
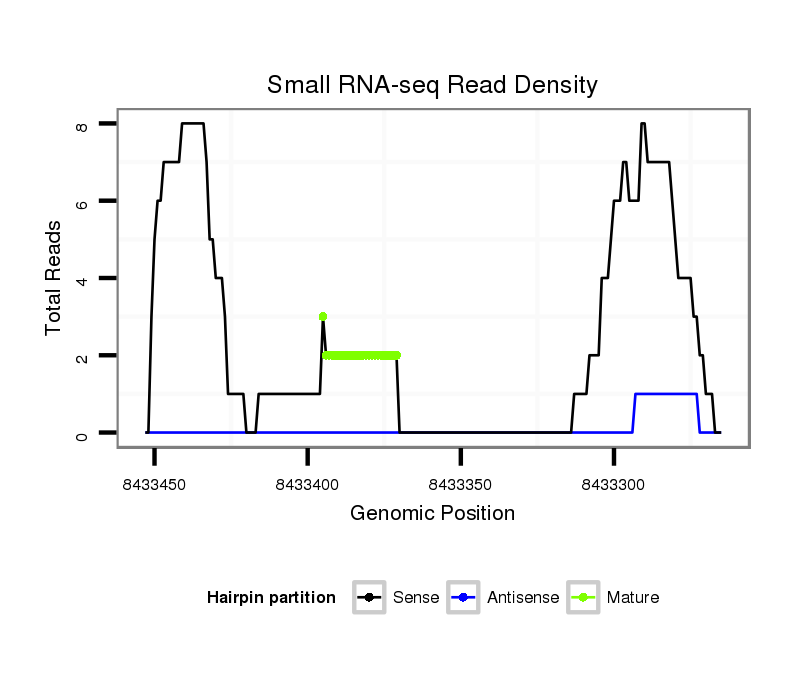


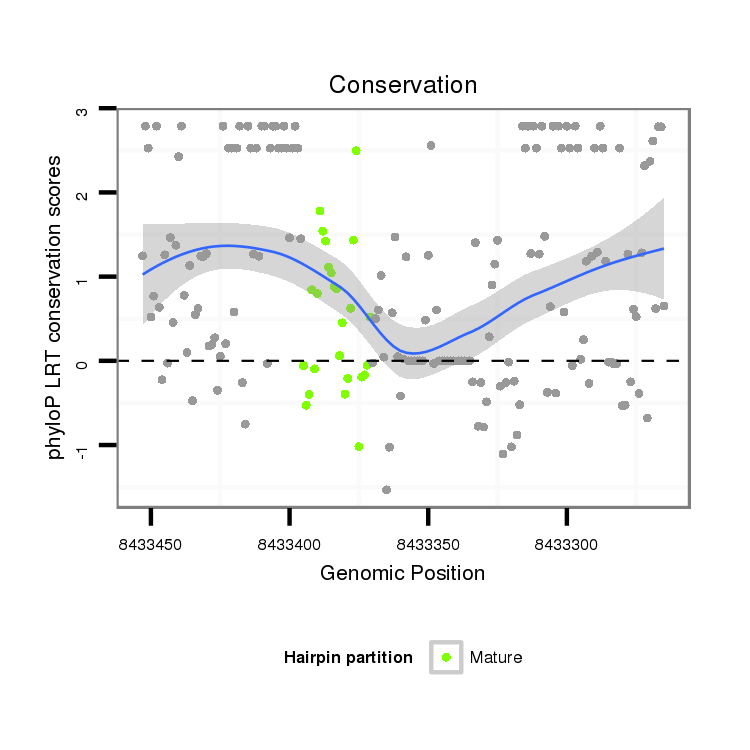
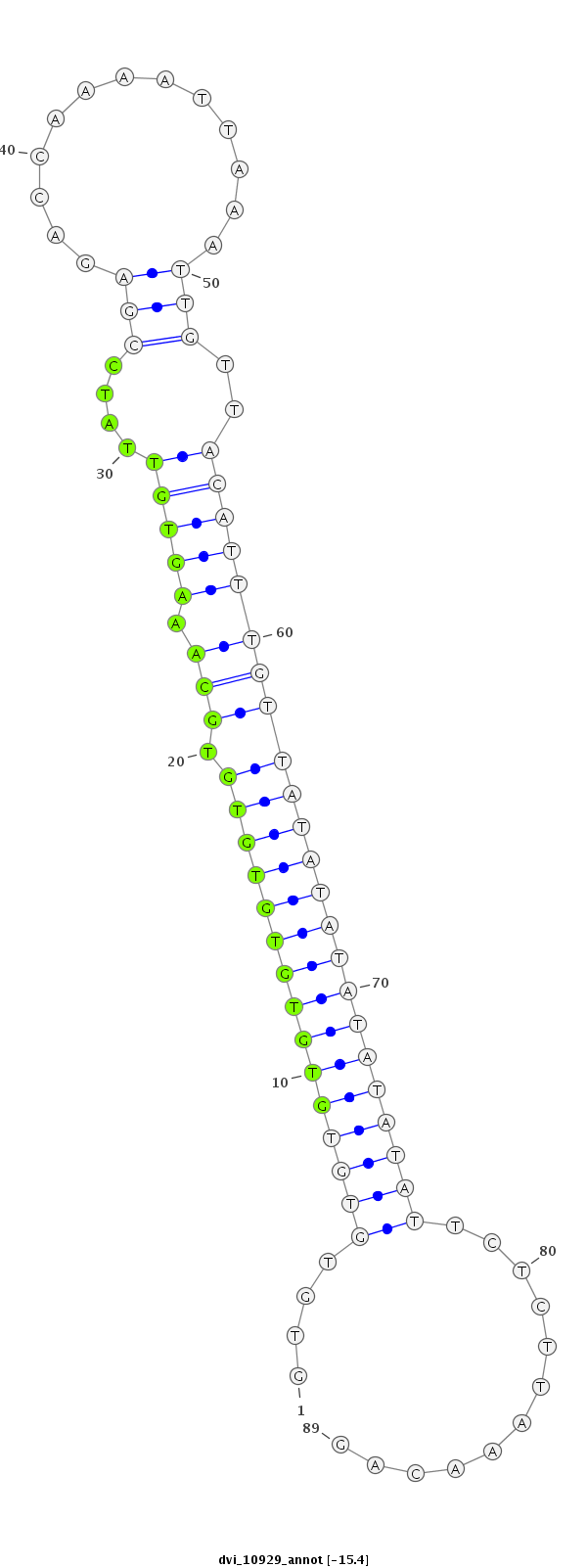
ID:dvi_10929 |
Coordinate:scaffold_12963:8433315-8433403 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -15.4 | -15.4 | -15.4 |
 |
 |
 |
CDS [Dvir\GJ10242-cds]; exon [dvir_GLEANR_1018:1]; CDS [Dvir\GJ10242-cds]; exon [dvir_GLEANR_1018:2]; intron [Dvir\GJ10242-in]
| Name | Class | Family | Strand |
| (CA)n | Simple_repeat | Simple_repeat | + |
| ##################################################-----------------------------------------------------------------------------------------################################################## GTCGATGACAAGACAGACGCTGCGGGCAATGCCGCAGATGGGCAAACAAGGTGTGTGTGTGTGTGTGTGTGCAAAGTGTTATCCGAGACCAAAATTAAATTGTTACATTTGTTATATATATATATATTCTCTTAAACAGTGAGCTATCTATCGACGAGCAGCGCCTGGCAGTCGGCAGCTGGTGAAATT **************************************************....(((((((((((((((.(((((.(((....(((.............)))..)))))))))))))))))))))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
M027 male body |
SRR060663 160_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
V116 male body |
M028 head |
M061 embryo |
SRR060655 9x160_testes_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060680 9xArg_testes_total |
SRR060681 Argx9_testes_total |
SRR060687 9_0-2h_embryos_total |
V047 embryo |
V053 head |
SRR060670 9_testes_total |
SRR060677 Argx9_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...GATGACAAGACAGACGCT........................................................................................................................................................................ | 18 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................GTGTGTGTGTGTGCAAAGTGTTATC.......................................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............ACAGACGCTGCGGGCAATGCC............................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ATGACAAGACAGACGCTGC...................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................AGCAGCGCCTGGCAGTCGGCAGC.......... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CGATGACAAGACAGACGCTGCGGG................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AAACAAGTGAGCTATCTATCGAC.................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................GCCTGGCAGTCGGCAGCTGGT...... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................ATCTATCGACGAGCAGCGC......................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ATCGACGAGCAGCGCCTGGCAGTCG............... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................ACGAGCAGCGCCTGGCAGTC................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CGATGACAAGACAGACGC......................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............AGACGCTGCGGGCAATGT............................................................................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CGATGACAAGACAGACGCTGCGGGC.................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................GCCTGGCAGTCGGCAGCTGGTGAA... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................ATGGGCAAACAAGGTGTGTGTG.................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ATCGACGAGCAGCGCCTGGCAGT................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GACAAGACAGACGCTGCGGGC.................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................GACGAGCAGCGCCTGGCAGTCGGCAGCTG........ | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GAGCTATCTATCGACGAG............................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .TCGATGACATTACAGACGC......................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......ACAAGGCAGAAGCTGCGGG................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................GCGATCTATCCATGAGCAG............................ | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................TTAAACAGTGCGATATCCAT...................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........GTCAGACGCAGCGGGCAA................................................................................................................................................................ | 18 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................CTGGCAGTGGTTAGCTGGT...... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CAGCTACTGTTCTGTCTGCGACGCCCGTTACGGCGTCTACCCGTTTGTTCCACACACACACACACACACACGTTTCACAATAGGCTCTGGTTTTAATTTAACAATGTAAACAATATATATATATATAAGAGAATTTGTCACTCGATAGATAGCTGCTCGTCGCGGACCGTCAGCCGTCGACCACTTTAA
**************************************************....(((((((((((((((.(((((.(((....(((.............)))..)))))))))))))))))))))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060678 9x140_testes_total |
SRR060670 9_testes_total |
SRR060689 160x9_testes_total |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
SRR060655 9x160_testes_total |
SRR060666 160_males_carcasses_total |
M028 head |
V116 male body |
SRR060671 9x160_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060658 140_ovaries_total |
SRR060663 160_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................TGACAAAAGGCACTGGTTTTAATTT.......................................................................................... | 25 | 3 | 2 | 102.00 | 204 | 54 | 44 | 38 | 20 | 26 | 12 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TTGACAAAAGGCACTGGTTTTAATTT.......................................................................................... | 26 | 3 | 2 | 75.00 | 150 | 35 | 20 | 43 | 26 | 8 | 8 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CAAAAGGCACTGGTTTTAATTT.......................................................................................... | 22 | 2 | 1 | 14.00 | 14 | 1 | 5 | 1 | 3 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GACAAAAGGCACTGGTTTTAATTT.......................................................................................... | 24 | 3 | 2 | 13.00 | 26 | 5 | 15 | 2 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ACAAAAGGCACTGGTTTTAATTT.......................................................................................... | 23 | 2 | 1 | 9.00 | 9 | 2 | 2 | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ACAAAAGGCACTGGTTTTAAT............................................................................................ | 21 | 2 | 2 | 4.50 | 9 | 0 | 2 | 0 | 0 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CAAAAGGCACTGGTTTTAAT............................................................................................ | 20 | 2 | 2 | 3.50 | 7 | 4 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ACAAAAGGCACTGGTTTTAATTTT......................................................................................... | 24 | 3 | 2 | 3.50 | 7 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TAGATAAGAGTAATTGTCAC................................................ | 20 | 3 | 14 | 3.14 | 44 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 11 | 4 | 5 | 5 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| .............................................................................CAAAAGGCACTGGTTTTAATTTT......................................................................................... | 23 | 3 | 2 | 3.00 | 6 | 2 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ACAAAAGGCACTGGTTTTAATT........................................................................................... | 22 | 2 | 2 | 1.50 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................CGCGGACCGTCAGCCGTCGAC........ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................AGGCACTGGTTTTAATTT.......................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................AAAAGGCACTGGTTTTAATTT.......................................................................................... | 21 | 2 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TGACAAAAGGCACTGGTTTTAATTTA......................................................................................... | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TTGACAATAGGCACTGGTTTTAAT............................................................................................ | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................AAAGGCACTGGTTTTAATTT.......................................................................................... | 20 | 2 | 3 | 0.67 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CAAAAGGCACTGGTTTTAA............................................................................................. | 19 | 2 | 3 | 0.67 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................AGATAAGAGTAATTGTCACTC.............................................. | 21 | 3 | 5 | 0.60 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TAGATAAGAGTAATTGTCACT............................................... | 21 | 3 | 7 | 0.57 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................ATAAGAGTAATTGTCACTC.............................................. | 19 | 2 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................AAAAGGCACTGGTTTTAAT............................................................................................ | 19 | 2 | 5 | 0.40 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TTGTTCCACACACACACACA............................................................................................................................. | 20 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ACAAAAGGCACTGGTTTTAA............................................................................................. | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................AAAAGGCACTGGTTTTAATT........................................................................................... | 20 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................AAAAGGCACTGGTTTTAATTTT......................................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TTCCACACACACACACACAC.......................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................ACAGACACACACACACGTTT.................................................................................................................. | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................TCCACACACACACACACAC.......................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:8433265-8433453 - | dvi_10929 | GTCGATGACAAGACAGACGCTGCGGGCAATGCCGCAGATGGGCAAACAAGGTGTGT-GTGTGTG------TGTGTGTG-------------------------CA--AAGTGTTATCCGA---------------------GA----------CCAAAATTAAATTGTTA-CATTTGTTATATATA---------TATAT-A--T-------T-------CT----CTTAAACAGTGAGCTATCTATCGACGAGCAGCGCCTGGCAGTCGG------CAGC---TG------GT------------------GAAATT |
| droMoj3 | scaffold_6500:1042341-1042535 - | GTCGATGGCAAAACAAACTCTGCGTGCAATGCCGCAGATGAGCAAACAAGGTGTGT-GTGTGTT------TG---------------------------ATTTTA--GTGTGCTTAAATT---------------------GCA---GCATATTTAATTTG------TAA-TA---------------------TTTGTCGT----------TAAT----------TTTGAATAGTGAGTTACTTATCGACGAGCAGCGCCTGCAGAGCCG------TCGT---TG------GTGGCAATGACAGCGTCGTCGAGATT | |
| droGri2 | scaffold_15252:11590183-11590369 - | GTCGGTGACAAAACAAACGCTGCGCTCGTTGCCGCAAATGGGCAAACAAGGTGTGT-GTGTGTG------TG---------------------------------GAAAGTG-----TGT------------------------GAAATA---TAAATTCT-------AATTA----------------------TAATCGTAATCCTT-----GT----------CTTCTTCAGTGAGCTATTTATCGACAAGCAGCGCCTGGCGGTCAG------CGGT---AG------GTGGCAACGATAGTGTTGTGGAGATT | |
| droWil2 | scf2_1100000004521:1804359-1804581 + | GTCGATGGCTAAACAAAGTCTGCGCTCTTTGCCGCAAATGGGCAAACAAGGTGTGT-GTGAAAATTGATTTGTG------------------------C---TTTTTGTGTGTGTGCCAAAAAAAAAAAAATAAACCAAATGCAAATGTA---ATAAAT--------------------------GTTCAAATGTTCTCTAT----------TATC----------TCTTATCAGTGAGCTATCTATCGACAAGCAGTGCCTGGCATGCTAGAGAGTCAGT---CG---------------GCTCTGTGGTGGAAATT | |
| dp5 | 4_group3:7873386-7873546 + | ATCCATGGCCAAACAATCCATGCGCGCACTGCCGCAGGTGGGCAAGCAAGGTGTGT-GTCTA------------------------------------ATCCTCCGCGAGTGTTAGC-------------------------------------CGAAAAT-------AATCA----------------------TCTCC-A----------TTTA----------CCAAACTAGTGAGCTACCTGTCGACGAGCAGCGCCTGGCGTGCAA------CAGT---TG------GTGGAAAC------------------ | |
| droPer2 | scaffold_1:9325006-9325170 + | ATCCATGGCCAAACAATCCATGCGCGCCCTGCCGCAGGTGGGCAAGCAAGGTGTGT-GTCTA------------------------------------ATCCTCCGCGAGTGTTAGC-------------------------------------CGAAAAT-------AATCA----------------------TCTCC-A----------TTTA----------CCAAACTAGTGAGCTACCTGTCGACGAGCAGCGCCTGGCGTGCAA------CAGT---TG------GTGGCAAC--------------GATT | |
| droAna3 | scaffold_12916:6681823-6682021 - | ATCCATGGCCAGACAATCCGCTAAAATGTTCCCCCAGGTGGGAAAACAAGGTGTGT-GCGAC-----------------------------------------TACCCAGTGACCATTGT---------------------GAA---GTGTTATTAATTAT-------AATTA----------------------TCTAT-T--TT------TTTATTATCTATTTTATTATTAGTGAGCTACCTGTCGACCAGTGCCTCCTGGAAGGCAT------CAAC---TG------GTGGCAACGACATGGTTGTGGAGATC | |
| droBip1 | scf7180000396568:130939-131124 + | ATCCATGGCCAGACAATCCGCTAAAATGTTTCCCCAGGTGGGAAAACAAGGTGTGT-GCGA-----------------------------------------TTATCCCGTGTCCATTGT---------------------GAA---GTGTTATTAATTAT-------AATTA----------------------TTCAT-A--TT------T-------------TTAAATTAGTGAGCTACCTGTCGACCAGTGCCTCCTGGAGGGCAT------CAAC---TG------GTGGCAACGACATGGTTGTGGAGATC | |
| droKik1 | scf7180000302472:1756709-1756903 - | ATCCATGGCCAGGCAATCGGCGCGAGCCCTTCCCCAGATGGGCAAGCAAGGTGTGT-GTTGTTG------TGTGTG------------------------C-TTA--GAGTGTTCCCTGT---------------------GC----------GCGGTGCT-------AACGA----------------------TTTAT-T--T-------T-------CGCCCTTCCGAGCAGTGAGCTACCTGTCGACGAGCAGCGCCTGGCGGGCAG------CCTCCGTGG------GCGGCAACGACATGGTGGTGGAGATT | |
| droFic1 | scf7180000454115:851651-851845 + | ATCCATTTCGA---AGTCAGCGCGAGTCCTTCCCCAAGTGGGCAAGCAAGGTGTGT-GTTCGTT------CGTG------------------------TGCTTTAGCGAGTGACCACAGT---------------------GC---AGTG---ACGATCCT-------AACGA----------------------TATATAT--TCCAT------------G----ATTTACTAGTGAGCTATCTATCCACGAGTGGAGCCTGGAAGGCGG------CAGC---TG------GAAACGGCGACATGGTGGTGGAGATC | |
| droEle1 | scf7180000491046:1513930-1514142 + | ATCCATGGCCAGACAAACGGCGCGAGCCCTTCCTCAGATGGGCAAGCAAGGTGTGTTGTGTGTG------TG----------CTTTAGCTTAAGCTTTAGCTTTAGCCAGTGTCCACTGT---------------------GC---AGTG---CTGATCCT-------AACGA----------------------TCTAT-T--T-------C-------CG----ATTTACCAGTAAGCTATCTCTCGACCAGTGGAGCCTGGAGGTCAG------CAGCCGCAG------GCGGTGGCGACATGGTGGTGGAGATC | |
| droRho1 | scf7180000779483:453265-453453 + | ATCCATAACCCGACAAACGGTGCGAGCCCTTCCCCAAGTGGGCAAGCAAGGTGTGT-GTGTG------------------------------------CGCTTTAGCGGTTGACCACTGT---------------------GC---AGTG---CTGATCCT-------AACGA----------------------TCTAT-T--T-------C-------CG----TTTTACCAGTGAGCTATCTCTCGACGAGCGGAGCCTGGAGGGCAG------CAGC---AG------GAGGTGGCGACATGGTGGTGGAGATC | |
| droBia1 | scf7180000301518:537324-537508 - | ATCCATGGCCAGACAAGCGGCGCGAGCCCTTCCCCAGATGGGCAAACAAGGTGGGT-GTGG--------------------------------------------GCGAGTGACCACTGT---------------------GCAGAGGTG---TTGACCCT-------AACGG----------------------TCTGTAT--T-------T-------CG----ATCTCCTAGTGAGCTGTCTTTCGACGAGCGGGGCCTGGAGGACGG------CCGC---CG------GGGGCGGCGATATGGTGGTGGAGATC | |
| droTak1 | scf7180000415876:329087-329278 - | ATCCATGGCTAGACAAACGGCGCGAGCCCTTCCCCAAGTGGGCAAGCAAGGTGTGT-GTGTGTG------------------------------------------CGAAGGACCACTGT------------------AGTGCAGAAGTG---CCGATCCT-------AACGG----------------------TCTAA-T--TTTAT-----GT----------TTTTACCAGTGAGCTATCTTTCGACGAGCGGCGCCTGGAGGGCAG------CAGC---CG------GCGGTGGCGACATGGTGGTGGAGATC | |
| droEug1 | scf7180000408936:16122-16307 - | ATCTATGGCCAGACAAACGGCGCGAGCCCTTCCCCAAGTGGGCAAGCAAGGTGTGT-GTGTG--------------------------------------CTTCAGCCAGTGA-CACTGT---------------------TTAGAAGTG---CTGATCCT-------AACGA----------------------TCTAT-T--T-------T-------CA-------ATTTAGTGAGCTATCTTTCTACGAGCGGAGCCTGGAGGGCAG------CAGC---CG------GAGGTGGTGACATGGTGGTGGAGATC | |
| dm3 | chr2L:7999118-7999316 + | ATCCGTGACCAGACAAACGGCGCGAGTCCTGCCCCAAATGGGCAAACAAGGTGTGT-GTGTGTG------TGTGTGTGT--------------------------GTGTGTGCGCACTGT---------------------GCTGGAGTG---GCGATGCT-------AACGT-------------------TCTTTCAA-T----------TTTT----------CATTAACAGTGAGCTATCTATCGACGAGTGGCGCTTGGCGGGCAA------CCGC---CA------GCGGTGGCGACATGGTGGTCGAGATC | |
| droSim2 | 2l:7736357-7736571 + | dsi_14046 | ATCCGTGACCAGACAAACGGCGCGAGTCCTGCCCCAAATGGGCAAGCAAGGTGTGT-GTGTGTG------TGTGTGTGTGTGCTTGTG----------CGCCCTAGCGAGTGCCCACTGT---------------------GCTGGAGTG---GCGATGCT-------AACGA-------------------TCCTTTCA-T----------TTTC----------CATTAACAGTGAGCTACCTATCGACGAGCAGCGCTTGGCGGGCAT------CCGC---CG------GCGGTGGCGACATGGTGGTCGAGATC |
| droSec2 | scaffold_3:3505828-3506034 + | dse_1174 | ATCCGTGACCAGACAAACGGCGCGAGTCCTGCCCCAAATGGGCAAGCAAGGTGTGT-GTGTGTG------TGTG--------CTTGTG----------CGCCTTAGCGAGTGCCCACTGT---------------------GCTGGAGTG---GCGATGCT-------AACGA-------------------TCCTTTCA-T----------TTTC----------CATTGACAGTGAGCTACCTATCGACGAGCAGCGCTTGGCGGGCAA------CCGC---CG------GCGGTGGCGACATGGTGGTCGAGATC |
| droYak3 | 2L:10632969-10633195 + | ATCCGTGAGCAGACAAACGGCGCGAGTCCTGCCCCAACTGGGCAAGCAAGGTGTGT-GTACATG------TGTGTGTGTGTGAGAGTG---------TCGCCTTAGCGAGTGCCTACTGT---------------------GCTGCAGTG---CCGATCCT-------AACGA----------------------ACTCC-T--CCCCCATTTTTA----------CATTGACAGTGAGCTACCTATCGACGAGCGGCGCTTGGAGAGCAT------CAGC---CGGCGGTGGCGGTGGCGACATGGTGGTCGAGATC | |
| droEre2 | scaffold_4929:16913886-16914066 + | ATCCGTGACCAGACAAACGGCGCGAGTCCTGCCCCAAGTGGGCAAGCAAGGTGTGT-GTGTGTG------T-----------------------------------------ACTACTGT---------------------GCTTCAGCG---CCGATCCT-------AACGA----------------------ACCCA-T----------TCTA----------CATTGACAGTGAGCTACCTATCGACGAGCGGCGCTTGGAGGGCCT------CAGC---CG------GCGGCAGCGACATGGTGGTCGAGATC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:04 AM