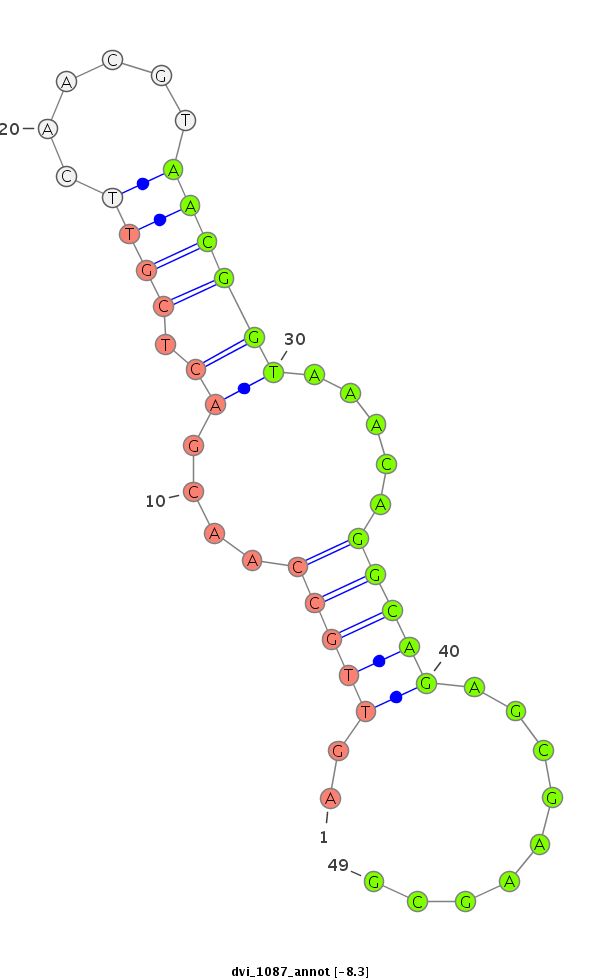
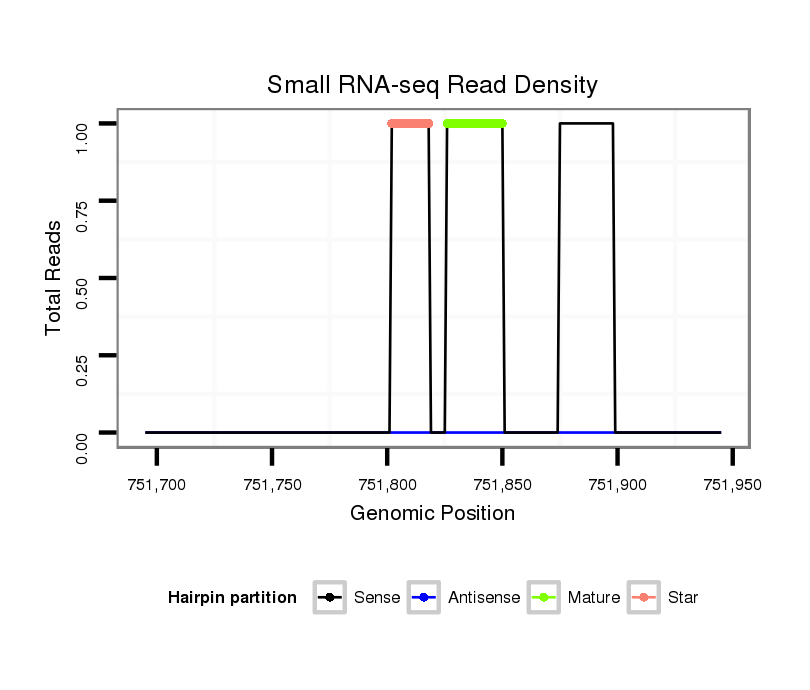
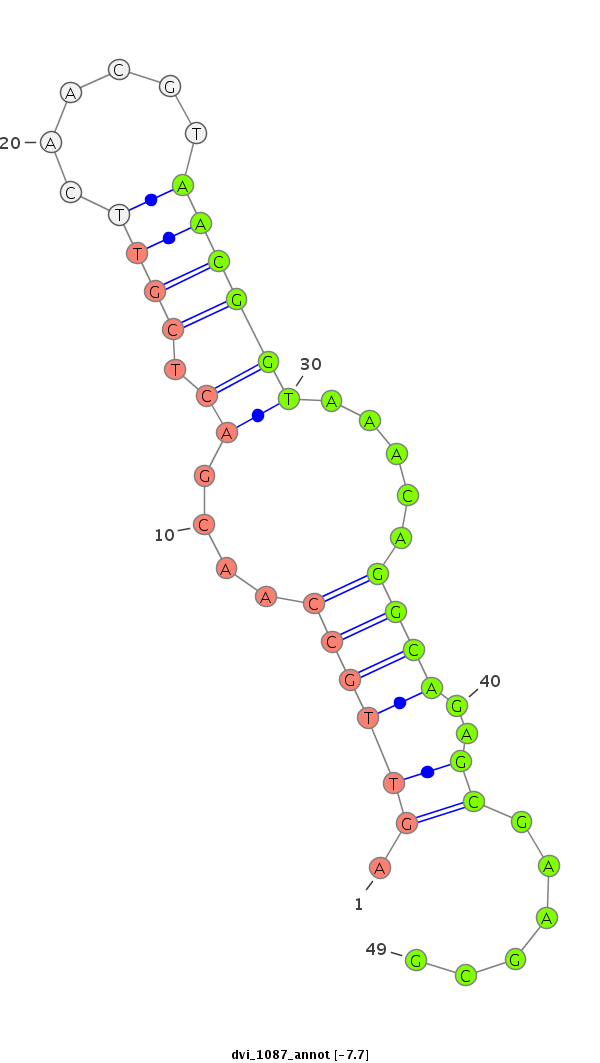
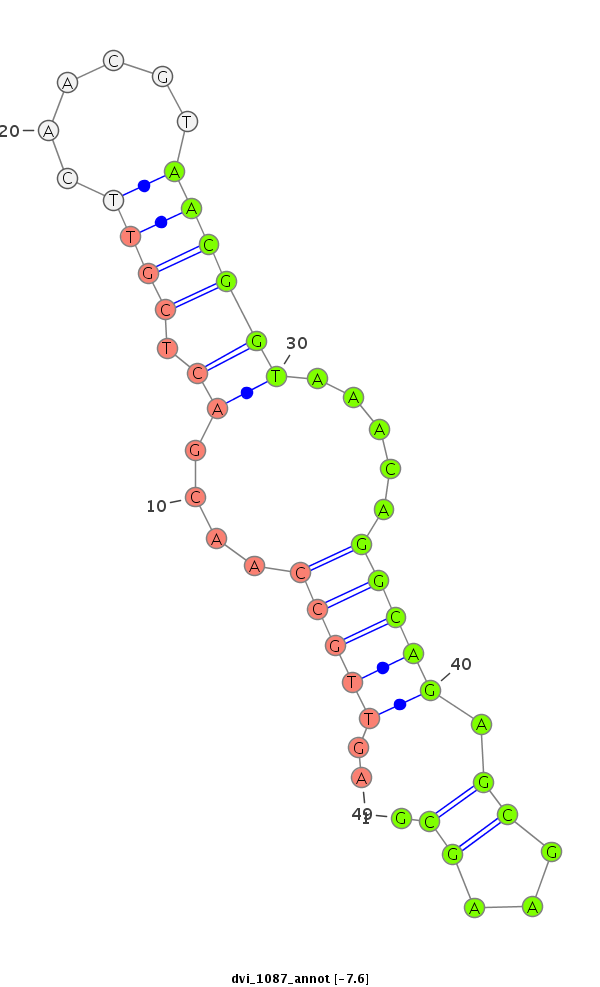
ID:dvi_1087 |
Coordinate:scaffold_12472:751745-751895 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
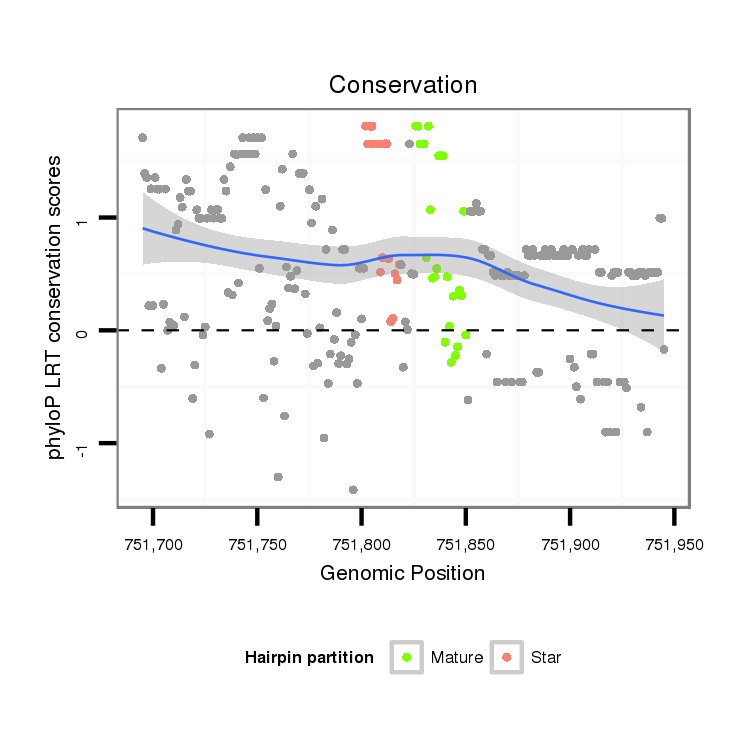
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -7.9 | -7.7 | -7.6 |
 |
 |
 |
exon [dvir_GLEANR_3364:1]; CDS [Dvir\GJ18568-cds]; intron [Dvir\GJ18568-in]
No Repeatable elements found
| --################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACATGTTGCAAGGCGGCATGTTGCGCTGCCGCTTGTTGCTGCCGCATGTGGTGAGTTTGGCGCAAACGAAACGCAGGCGTCGGCACACTGGCCCCTTTGGCTCGGCCAGTTGCCAACGACTCGTTCAACGTAACGGTAAACAGGCAGAGCGAAGCGCGCCAGCAAACGACGACGTTGGCGCCTGACGTCGGCGTCGTGGGCGTCGTTGTCGTCGTATACATCCCCGAGACGCTAACCGTGACCATAAACGT ***********************************************************************************************************..(((((....((.((((......)))))).....))))).........*********************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060669 160x9_females_carcasses_total |
SRR060681 Argx9_testes_total |
V116 male body |
SRR060663 160_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060665 9_females_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060673 9_ovaries_total |
SRR1106730 embryo_16-30h |
SRR060659 Argentina_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060680 9xArg_testes_total |
SRR060666 160_males_carcasses_total |
SRR060687 9_0-2h_embryos_total |
M061 embryo |
M028 head |
SRR060683 160_testes_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................TCCCTGAGACCCTAACTGTGA.......... | 21 | 3 | 1 | 9.00 | 9 | 1 | 2 | 2 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................CATGTGGGGAGTTTGGCT............................................................................................................................................................................................. | 18 | 2 | 7 | 1.14 | 8 | 4 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AACGGTAAACAGGCAGAGCGAAGCG............................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................ATGTTGGTGGCGCATGTGGTGA..................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................CCTGACGTCGGCGTCGTGGGCGTC............................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................AGTTGCCAACGACTCGT............................................................................................................................... | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................CAGGTGGGGAGTTTGGCGC............................................................................................................................................................................................ | 19 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................GCAGGTGGGGAGTTTGGC.............................................................................................................................................................................................. | 18 | 2 | 7 | 0.43 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TCCCTGAGACCCTAACTGTG........... | 20 | 3 | 5 | 0.40 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................CAGGTGGGGAGTTTGGCG............................................................................................................................................................................................. | 18 | 2 | 7 | 0.29 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TCATGTGGGGAGTTTGGC.............................................................................................................................................................................................. | 18 | 2 | 7 | 0.29 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TATGGTGAGTTTGGCGCACATG....................................................................................................................................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGAGTTTGGGGTAAACGAA..................................................................................................................................................................................... | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....TTTCAAGGCGGATTGTTGCGC................................................................................................................................................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................AAAGGAAGCGCAGGAGTCGGC....................................................................................................................................................................... | 21 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................CTTTGCCTCGGCCAGTTGACT........................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................CAGGTGGTGAGTTTGGCT............................................................................................................................................................................................. | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................GTTGGTGGCGCATGTGGT....................................................................................................................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGGGGGGAGTATGGCGCAA.......................................................................................................................................................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................CTTGTTCAAGGTACCGGTA................................................................................................................. | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TAACGATAAACAGGGTGAG...................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................AGGTGGGGAGTTTGGCACA........................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................AAAGGAAACGCAGGAGTCGGC....................................................................................................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................AGAGGAAACGCAGGAGTCGGC....................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................AAGGGAAACGCAGGAGTCGGC....................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
TGTACAACGTTCCGCCGTACAACGCGACGGCGAACAACGACGGCGTACACCACTCAAACCGCGTTTGCTTTGCGTCCGCAGCCGTGTGACCGGGGAAACCGAGCCGGTCAACGGTTGCTGAGCAAGTTGCATTGCCATTTGTCCGTCTCGCTTCGCGCGGTCGTTTGCTGCTGCAACCGCGGACTGCAGCCGCAGCACCCGCAGCAACAGCAGCATATGTAGGGGCTCTGCGATTGGCACTGGTATTTGCA
***********************************************************************************************..(((((....((.((((......)))))).....))))).........*********************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060662 9x160_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
M047 female body |
V047 embryo |
SRR060657 140_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060671 9x160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TTGCTGCTGCAATCGTGG..................................................................... | 18 | 2 | 5 | 0.40 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................CTGAAATCGCGGCCTGCAG.............................................................. | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................TCCCTTCGCGCGATGGTT...................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ...........................................................................................CGGGAAGCCTAGCCGGTCAA............................................................................................................................................ | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................GCAGCCGCGTCACCCGCA................................................ | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................GGAACAACGACGGCGTCCA.......................................................................................................................................................................................................... | 19 | 2 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................CAACGGGTGCTGGGCCAG............................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................TCGCGTGGCCCGGTCGTTT..................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12472:751695-751945 + | dvi_1087 | ACATGT-TGCAAGGCGGCATGTTGC--------GCTGCCGCTTGTTGCTGCCGCA------TGTGGTGAGTTTG-------------------------------------------GCG--CAA--------ACGAA-----------ACGCAGGCGTCGGCACACTG------------G------CCC-CTTTG----GCTCGGCCAGTTGCCAACGACTCGTTCAACGTAACGGTAA-A--CAGGCAGAGCGAAGCGCGCCAGCAAACGACGACGTTGGCGCCTGACGTCGGCG----TCGTGGGCG--TCGTTGTCGTCGTATACATCCCCGAGACGCTAA---CCGTGACCA----------TAAACGT |
| droMoj3 | scaffold_6308:2676924-2677126 - | ACATGT----------------TGCGC-TGTGGCATGCCTCGTGTTGCTGCCGC------CTGTGGTGAGTTTC-------------------------------------------GCG--CAG--------CCGAA-----------ACGCAGGCGCC-ACGCAGTT------------G------CCC-CATTGGCGGCGGCTGCCAGTTGCCAGCGATTCGTTCAACGTAACGGTAACGGCTAGGCAGAGCGAAGCGCGCCAGCAAACG----------------ACGTCAACG----TCGTGGGCG--TCGCTTGCCTCGTATA-------------------------------------------CGT | |
| droGri2 | scaffold_15203:3852528-3852795 - | AC-----TGCCAG-CAACATGTTGCAG--------TGCGGCATGTTGCTGCCACA------TGTGGTGAGTTTA-------------------------------------------GCGTACAG--------GCGCA-----------ACGCAGGCGCCGGC--ACTGGT----------G-------GCCATTTG----GGCCGACTAGTTGCCAACGGCTCGTTCAACGTAACGGTAACG--CAGGCAGACCGAAGCGGGCCAGCAAGCGACAACGCTGACGCTCGACGTCAACGCTTGTCGTGGGCGTGTCGTTGTCGTCGTGCATATTGTGGACATATAAACGTCCGTTACGAAGCAAATTGTTAAACGC | |
| dp5 | XL_group3a:556424-556610 + | A------------------TGTTGCAGTTGGAAGCTGCCGCATGTTGCTGCCACAAGTTGCTGTGGTGAGTTTTCATGC----TGCCACGCGACGTCCACGCAACATGTTGCTGC--------TGCCACACC---GAAGCATTCAAACGACGGG---------------GCCGCCT--------------------C----CGCTAGTCAGTTGCCCACGATAACGGTAACTTAACGGTAA-G--CAGGCAAGGACGAG-----------------------------------------------------------------------------------------------------------------CGC | |
| droPer2 | scaffold_39:104479-104665 + | A------------------TGTTGCAGTTGGAAGCTGCCGCATGTTGCTGCCACAAGTTGCTGTGGTGAGTTTTCATGC----TGCCACGCGACGTCCACGCAACATGTTGCTGC--------TGCCACACC---GAAGCATTCGAACGACGGG---------------GCCGCCT--------------------C----CGCTAGTCAGTTGCCCACGATAACGGTAGCTTAACGGTAA-G--CAGGCAAGGACGAG-----------------------------------------------------------------------------------------------------------------CGC | |
| droKik1 | scf7180000302685:581826-581999 + | ACACGTTTGCCGG-CAACATGCTGC----------------G------TGCCACAAGTTGCCGTGGTGAGTCTGC--GCCAGTTGCGCCACGACGCCTACGCAACATGTTGCTGC--------CG------------A-----------ATGGAGGCGT---------------CTCCGGCAGCCAGTCGTC-C---------ATCGCCAGTTGCCCACGACGCGTTATACGCAACGGAA--G--CAGGCGAAGCGGAG-------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453829:42246-42391 + | ACATGC-TGCCGG-TAA-----------------------------------ACAAGTTGCCGTGGTGAGTTTGC--GC----CGCTTGGCGTCGCCCACGCAACATGTTT-----GGGGCATCGCCTCAGT---GGA-----------GCGGAGGCATC--------AGTC-----------------GCC-C--T----CGGCCGTCAGTTGCCCACGACGCGTTCCTCGCAACGGTA--------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779985:1411279-1411336 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTCAGTTGCCCACGACGCGTTCCTCGCAACGGTA--G--CAGGCGAGCCATCTCCTGCCAGC----------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409458:150997-151155 + | ACATGT-TGCTGG-------------------------------------ACACATGTTGCCGTGGTGAGTTTGC--GC----CGGTCCTCATCGCCCACGCAACATGTT----CTGGGACATTA--TAAATATCGTT-----------GCGGAGGCGTCGACGTAG--------TC----CG-------------C----CAGTCGTCAGTTGCCCACGACGCGTTACTCGCAACGGTA--G--CTGGCCAG-------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/19/2015 at 10:58 AM