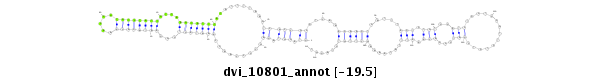
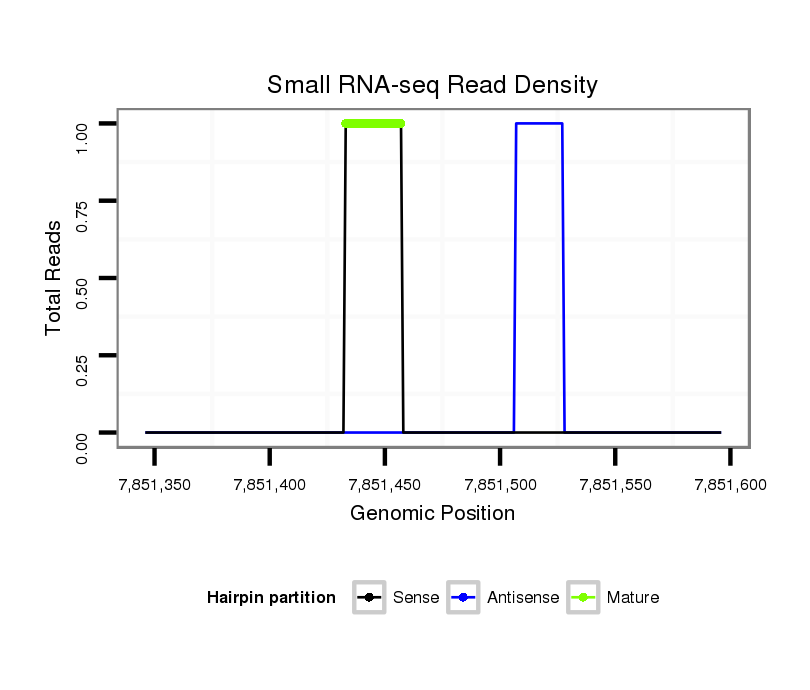
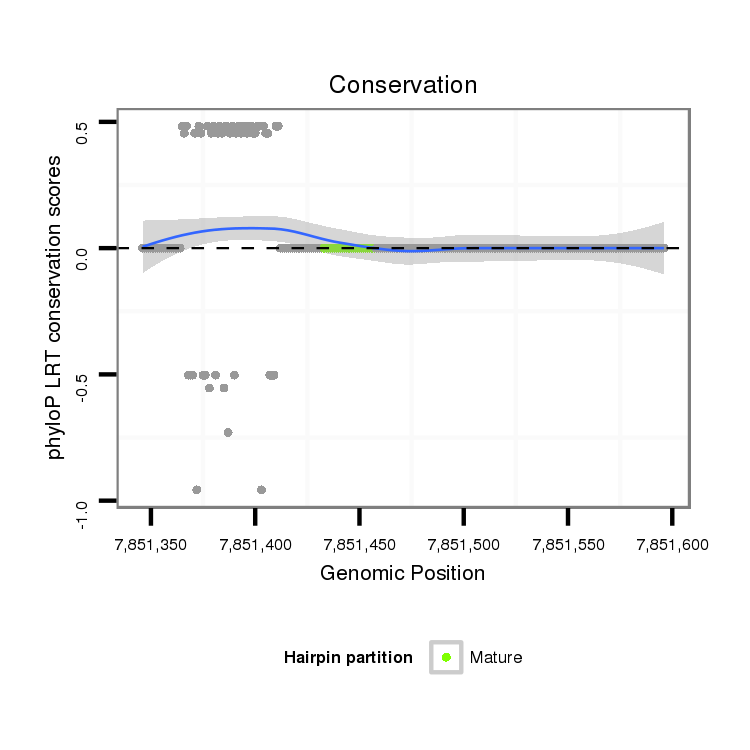
ID:dvi_10801 |
Coordinate:scaffold_12963:7851396-7851546 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -19.4 | -19.2 | -19.1 |
 |
 |
CDS [Dvir\GJ17159-cds]; exon [dvir_GLEANR_1770:1]; intron [Dvir\GJ17159-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CAAAACCATCCGAGTCACCAGAGACCGACCATACTGGTCATCGTCGTCAAGTAGGTAGAGCCCCAACCAAATACTCGGAATAGACAAACCGTGTCTATTGTTAAGTATTTATATTTTAAGTCACCGCGTCAAACAAGTCATTTAGTCTGTCCATCTGCTCCGTTCTCTCGATCCCACAAATTTATAAAAGCTTGTCTAGGTATCTTGGTATTTCTGTGCCAGCTTCTAGCCACGAGATTTCCGTAAACTAA **************************************************((.(((.((.........(((((((...((((((((.....))))))))....)))))))..........))))))).....((((((.....(((.(((..(((..............)))...))).))).......))))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
V053 head |
SRR060672 9x160_females_carcasses_total |
SRR060683 160_testes_total |
M028 head |
SRR060659 Argentina_testes_total |
SRR060660 Argentina_ovaries_total |
V116 male body |
SRR060665 9_females_carcasses_total |
SRR1106726 embryo_14-16h |
M047 female body |
M027 male body |
SRR060654 160x9_ovaries_total |
SRR060656 9x160_ovaries_total |
SRR060670 9_testes_total |
SRR060673 9_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................TCTCGATCCCGCGAAATTA................................................................... | 19 | 3 | 12 | 1.33 | 16 | 0 | 0 | 0 | 3 | 0 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| .......................................................................................ACCGTGTCTATTGTTAAGTATTTAT........................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................TAAGTCACCGAGACAAACAGGT................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................CTATCGATCCCGCAATTTTAT.................................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CCATACTGATCGTCGTCGT............................................................................................................................................................................................................ | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................GGGGCATCGTGGTCAAGTA...................................................................................................................................................................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................GGTAGATCTGTGCCAGCTCC......................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................AGAGTCCGACGATACTGCT..................................................................................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CGACCAAAAACTCGGAAAAG........................................................................................................................................................................ | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................GATCGTGGTCAAGAAGGTA.................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................ATAAAAGCTACTCTAGGGA................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CAGTTGGTAGAGCCCCAAA........................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTTTTGGTAGGCTCAGTGGTCTCTGGCTGGTATGACCAGTAGCAGCAGTTCATCCATCTCGGGGTTGGTTTATGAGCCTTATCTGTTTGGCACAGATAACAATTCATAAATATAAAATTCAGTGGCGCAGTTTGTTCAGTAAATCAGACAGGTAGACGAGGCAAGAGAGCTAGGGTGTTTAAATATTTTCGAACAGATCCATAGAACCATAAAGACACGGTCGAAGATCGGTGCTCTAAAGGCATTTGATT
**************************************************((.(((.((.........(((((((...((((((((.....))))))))....)))))))..........))))))).....((((((.....(((.(((..(((..............)))...))).))).......))))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
M061 embryo |
SRR060673 9_ovaries_total |
V047 embryo |
SRR060657 140_testes_total |
SRR060665 9_females_carcasses_total |
SRR060670 9_testes_total |
SRR060674 9x140_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060663 160_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................CAAGAGAGCTAGGGTGTTTAA..................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................AGAGCTAGGGCGCTTTAAT................................................................... | 19 | 3 | 12 | 0.42 | 5 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| ..........................CTGGTCTGAACAGAAGCAG.............................................................................................................................................................................................................. | 19 | 3 | 11 | 0.18 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AATCGGATAGGTAGACGA............................................................................................ | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................CTCTGGCCTCTATGACCAG.................................................................................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................CAAGTAGACGTGGCATGAG.................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .TTTAGGTAGGCTCTGTAGTC...................................................................................................................................................................................................................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..TTTGGTAGGTTCAGTAGTCG..................................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:7851346-7851596 + | dvi_10801 | CAAAACCATCCGAGTCACCAGAGACCGACCATACTGGTCATCGTCGTCAAGTAGGTAGAGCCCCAACCAAATACTCGGAATAGACAAACCGTGTCTATTGTTAAGTATTTATATTTTAAGTCACCGCGTCAAACAAGTCATTTAGTCTGTCCATCTGCTCCGTTCTCTCGATCCCACAAATTTATAAAAGCTTGTCTAGGTATCTTGGTATTTCTGTGCCAGCTTCTAGCCACGAGATTTCCGTAAACTAA |
| droMoj3 | scaffold_6500:3000402-3000448 - | -------------------AGAAGTCCACTGTTCTAGTCTTAGTTGTCAAGTAGGTACAGCTTTAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 10:09 PM