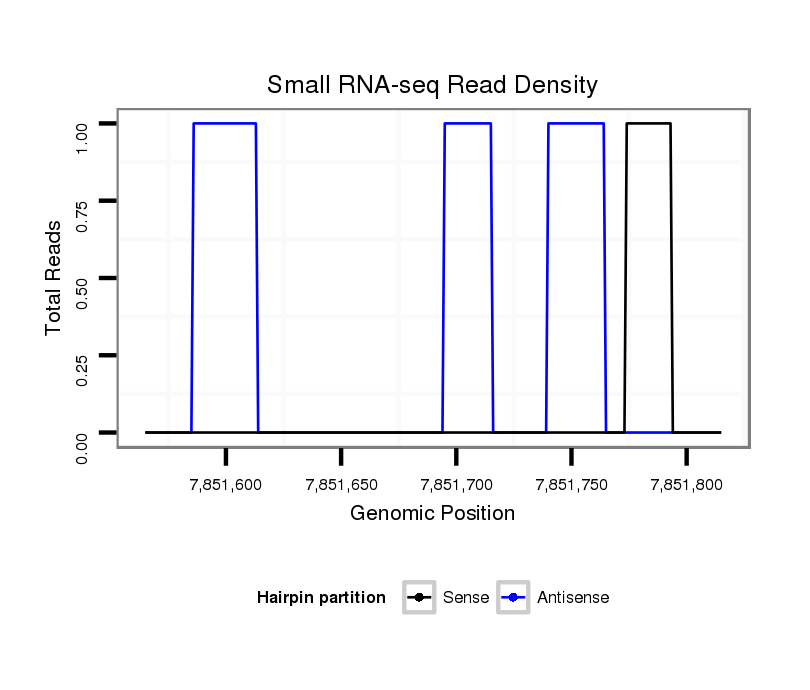
ID:dvi_10798 |
Coordinate:scaffold_12963:7851615-7851765 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
No conservation details. |
exon [dvir_GLEANR_1770:2]; CDS [Dvir\GJ17159-cds]; intron [Dvir\GJ17159-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CAGCTTCTAGCCACGAGATTTCCGTAAACTAAATCTTATAAACTGGAGCTTATGATAGTTATAGAATAAGCCAAGTAGCTTGTTATGTGTGTCTGTCTGTTGAGATCTCTGAATACAGGAAATTCTCTGGTCGGAGCCCTTTCGACTAGGCCGCTTTACTTTGCCCTATAGCTAAGTATATTTGATACATTCCCCATTTAGCAACAAAAATCTAGCTGGAGCCGCTTTAAATAATGGCTATAAGCCCGTGG **************************************************.....((((((((((((((((......)))))))..(((.(((((...(((((((.((((((...((((......)))).))))))...)))))))))))))))...........))))))))).((((.....)))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
V047 embryo |
SRR060664 9_males_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................ATCTAGCTGGAGCCGCTTTA...................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................ATATGATAGTTACAGAATAA...................................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................TCTGTTTAGATCTCGGAGTA........................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................AATACTTTGGTTGGAGCCCT............................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................CATTGCCCGGTAGCTAAGT.......................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TTGGAGCCGCTTTAAAAAA................. | 19 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................GAATATTTGATCCATTACCC........................................................ | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................TATTGGGATCTCTGAATGC....................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GTCGAAGATCGGTGCTCTAAAGGCATTTGATTTAGAATATTTGACCTCGAATACTATCAATATCTTATTCGGTTCATCGAACAATACACACAGACAGACAACTCTAGAGACTTATGTCCTTTAAGAGACCAGCCTCGGGAAAGCTGATCCGGCGAAATGAAACGGGATATCGATTCATATAAACTATGTAAGGGGTAAATCGTTGTTTTTAGATCGACCTCGGCGAAATTTATTACCGATATTCGGGCACC
**************************************************.....((((((((((((((((......)))))))..(((.(((((...(((((((.((((((...((((......)))).))))))...)))))))))))))))...........))))))))).((((.....)))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060688 160_ovaries_total |
V116 male body |
V053 head |
M027 male body |
SRR060681 Argx9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................CATATAAACTATGTAAGGGGTAAAT................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................AGCCTCGGGAAAGCTGATCCG.................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................GGCATTTGATTTAGAATATTTGACCTCG.......................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TATTGAGTTCATCCAACAATA..................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................CATTGGATTTAGAATCTT.................................................................................................................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................AAAGGTATTTGACTTAGA....................................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:7851565-7851815 + | dvi_10798 | CAGCTTCTAGCCACGAGATTTCCGTAAACTAAATCTTATAAACTGGAGCTTATGATAGTTATAGAATAAGCCAAGTAGCTTGTTATGTGTGTCTGTCTGTTGAGATCTCTGAATACAGGAAATTCTCTGGTCGGAGCCCTTTCGACTAGGCCGCTTTACTTTGCCCTATAGCTAAGTATATTTGATACATTCCCCATTTAGCAACAAAAATCTAGCTGGAGCCGCTTTAAATAATGGCTATAAGCCCGTGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/16/2015 at 10:10 PM