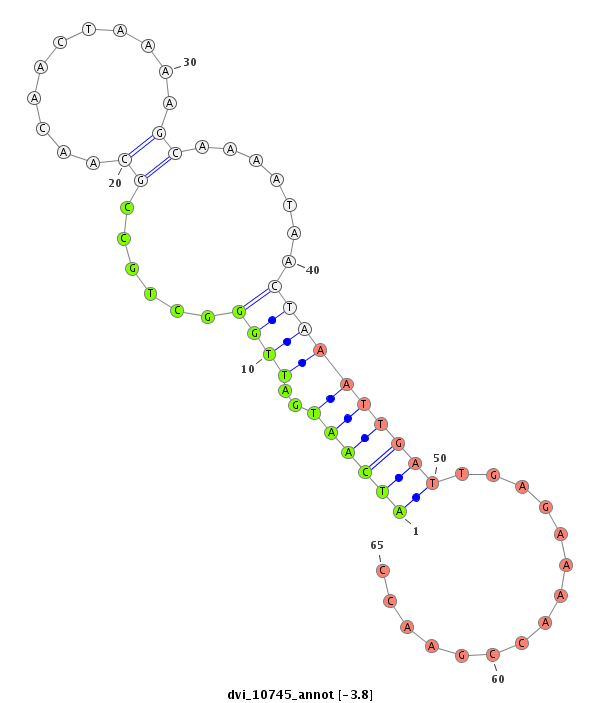
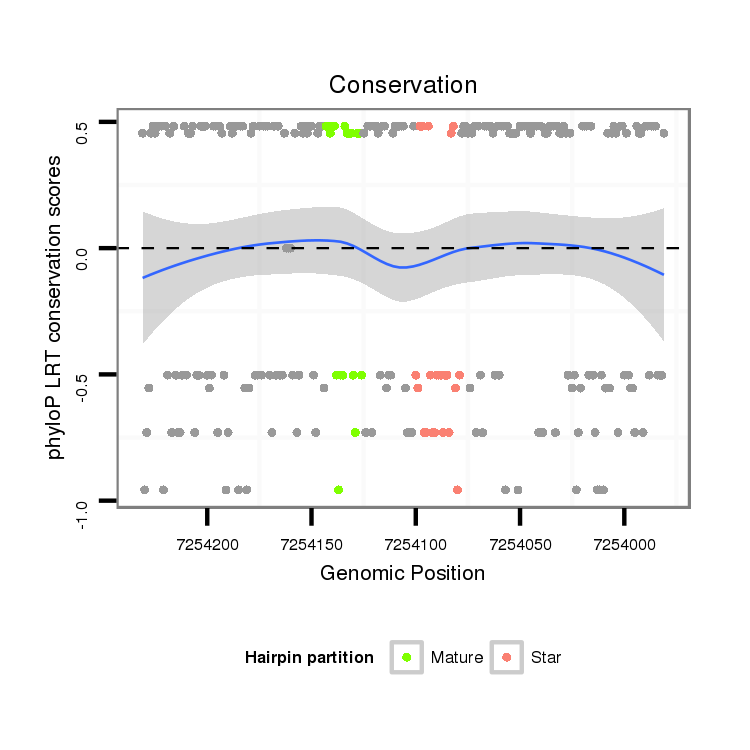
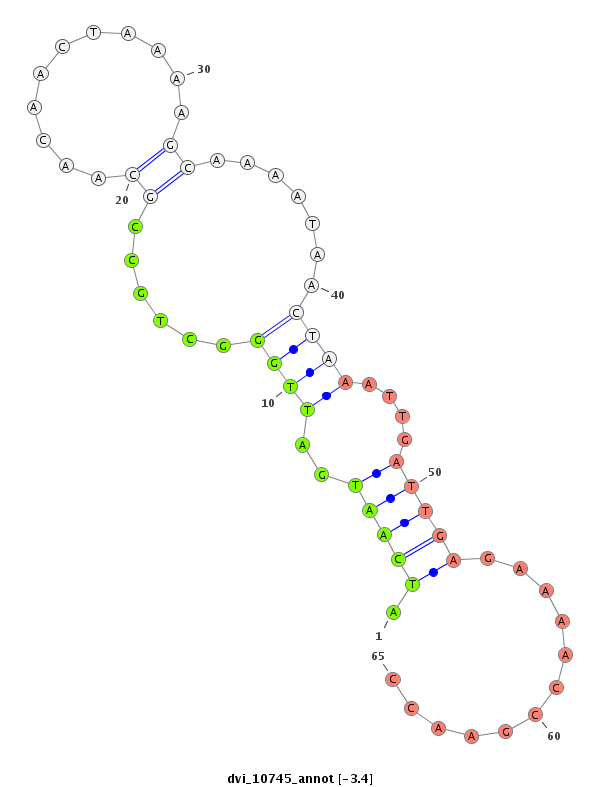
ID:dvi_10745 |
Coordinate:scaffold_12963:7254031-7254181 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
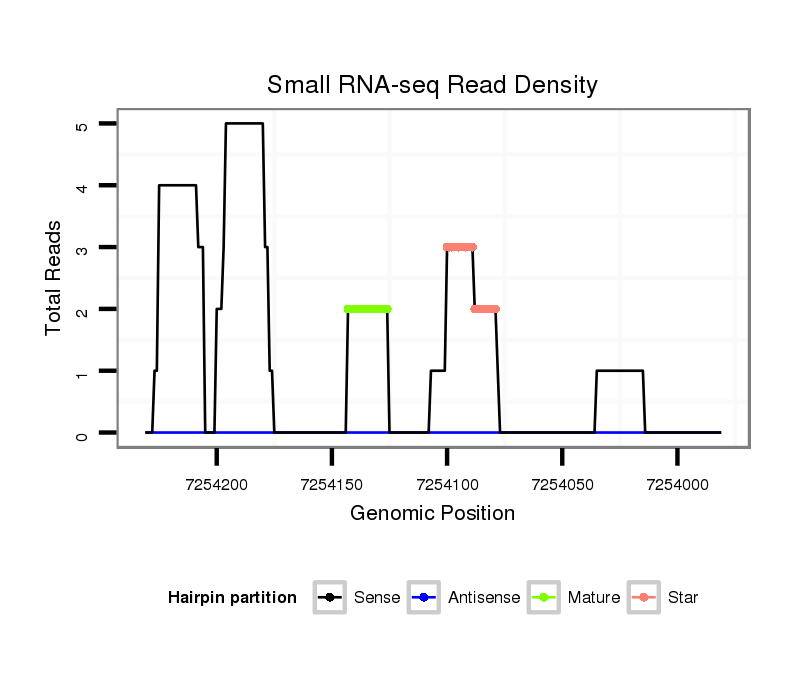
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -3.4 | -3.4 | -3.3 |
 |
 |
 |
exon [dvir_GLEANR_1072:1]; CDS [Dvir\GJ10820-cds]; intron [Dvir\GJ10820-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCCAGAGTATGAGGAAAAGCTGCGTAGAATTCACAAGTCTCCAGTTGAAAGTGTGAACTTTGCAGACTCCGCGGCGACTGTTTCCAAAATCAATGATTGGGCTGCCGCAACAACTAAAAGCAAAATAACTAAATTGATTGAGAAAACCGAACCCAACTCAAAGATATTGTTTATGAGCGCGATTTACTTTCACGGATTGTGGAGGCTCGATTTTGATAGCGCACACACTAAGCGTTCACCCTTCTATGTAC ****************************************************************************************((((((..((((......((...........)).......))))))))))...............************************************************************************************************** |
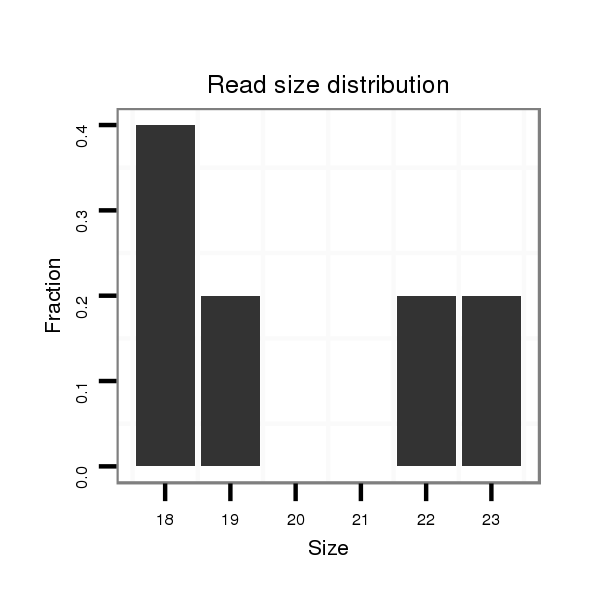
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060655 9x160_testes_total |
SRR060659 Argentina_testes_total |
SRR060679 140x9_testes_total |
SRR060683 160_testes_total |
V116 male body |
SRR060657 140_testes_total |
SRR060689 160x9_testes_total |
M027 male body |
SRR060661 160x9_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
SRR060666 160_males_carcasses_total |
V047 embryo |
M028 head |
SRR060663 160_0-2h_embryos_total |
SRR060673 9_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......GTATGAGGAAAAGCTGCGTA................................................................................................................................................................................................................................. | 20 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AGTCTCCAGTTGAAAGTGT..................................................................................................................................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................CACAAGTCTCCAGTTGAAAGT....................................................................................................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................ATCAATGATTGGGCTGCC................................................................................................................................................. | 18 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TTACCGGATTGTGGAGGCTC........................................... | 20 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................AAGTCTCCAGTTGAAAGTGTGA................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................AAACTCAAGGATTGGTCTGCCGC............................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TTGTGGAGGCTCGATTTTGAT.................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AATTGATTGAGAAAACCGAACC.................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AATTGATTGAGAAAACCGAACCC................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................ATAACTAAATTGATTGAGA............................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GAGTATGAGGAAAAGCTGC.................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................AAATGGGAACTTTGAAGACT....................................................................................................................................................................................... | 20 | 3 | 18 | 0.78 | 14 | 10 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| ......GTATCAGGAAAAACTGCGTA................................................................................................................................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................AAATGAGAACTTTGCAGACT....................................................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CAAGTGGGAACTTTGAAGACT....................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................AACTCAAGGATTGGTCTGCCG................................................................................................................................................ | 21 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........CAGGAAAAACTGCGTAGAATT............................................................................................................................................................................................................................ | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................AAATGGGAACTTTGCAGACT....................................................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TTACCGGATTGTGGAGGCT............................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................AACTCAAGGATTGGTCTGCC................................................................................................................................................. | 20 | 3 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AGTCTCAACTTTGATAGCGC............................. | 20 | 3 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................AAATGTGAACTTTGAAGAC........................................................................................................................................................................................ | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................CTGGGAACTTTGCAGACT....................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TTAAGGGATTTTGGAGGCTC........................................... | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AAAATGAGAACTTTGAAGACT....................................................................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TTGGCAGAATCCGCGGGGA.............................................................................................................................................................................. | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................AAATGAGAACTTTGCAGA......................................................................................................................................................................................... | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ............................................................................................................................ATTACTAAATTGATAGAGAAT.......................................................................................................... | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................AATGAGAACTTTGAAGACTC...................................................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................AAGTGAGAACTTTGAAGA......................................................................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GATACTAAGCGTTCACCCA......... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................AGTCAATGGTTGGGTTGCC................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
CGGTCTCATACTCCTTTTCGACGCATCTTAAGTGTTCAGAGGTCAACTTTCACACTTGAAACGTCTGAGGCGCCGCTGACAAAGGTTTTAGTTACTAACCCGACGGCGTTGTTGATTTTCGTTTTATTGATTTAACTAACTCTTTTGGCTTGGGTTGAGTTTCTATAACAAATACTCGCGCTAAATGAAAGTGCCTAACACCTCCGAGCTAAAACTATCGCGTGTGTGATTCGCAAGTGGGAAGATACATG
**************************************************************************************************((((((..((((......((...........)).......))))))))))...............**************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060668 160x9_males_carcasses_total |
V116 male body |
V047 embryo |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
SRR060678 9x140_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................CGCGCTAAATAAAAGTGTCTC...................................................... | 21 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................AGAGGTCCACTTTCGCAC.................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................GTGATCCGCAAGAGGGTAG....... | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................GCAAGTTGGAAGACACA.. | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................AAAACCATCGAGTGTGTGAC..................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................CTGACCAAGGTTATAGTG.............................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................TGGCAAAGGTTTTAATTA............................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:7253981-7254231 - | dvi_10745 | GCCAGAGTATGAGGAAAAGCTGCGTAGAATTCACAAGTCTCCAGTTGAAAGTGTGAACTTTGCAGACTCCGCGGCGACTGTTTCCAAAATCAATGATTGGGCTGCCGCAACAACTAAAAGCAAAATAACTAAATTGATTGAGAAAACCGAACCCAACTCAAAGATATTGTTTATGAGCGCGATTTACTTTCACGGATTGTGGAGGCTCGATTTTGATAGCGCACACACTAAGCGTTCACCCTTCTATGTAC |
| droMoj3 | scaffold_6680:14618774-14619021 - | GGATGAGTATCAAGCAGCTTTACGTCAGATTTTTAATTCCGAAGTTCAATCAGTAGATTTTAAAAATCC---AGAAACTGTTCACAATATCAACCGCTGGGTGGCTGAAAAAACCAATGACAAAATTCAGAGTTTTCTCTCAGGCGTAGATGTCAACACACAACTATTGCTCATCAGCGCCATTTACTTTAAAGGATTTTGGAGACATCCATTTAACCCGAGTCTCACGAAACAAAAACCATTTTATACGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 09:18 PM