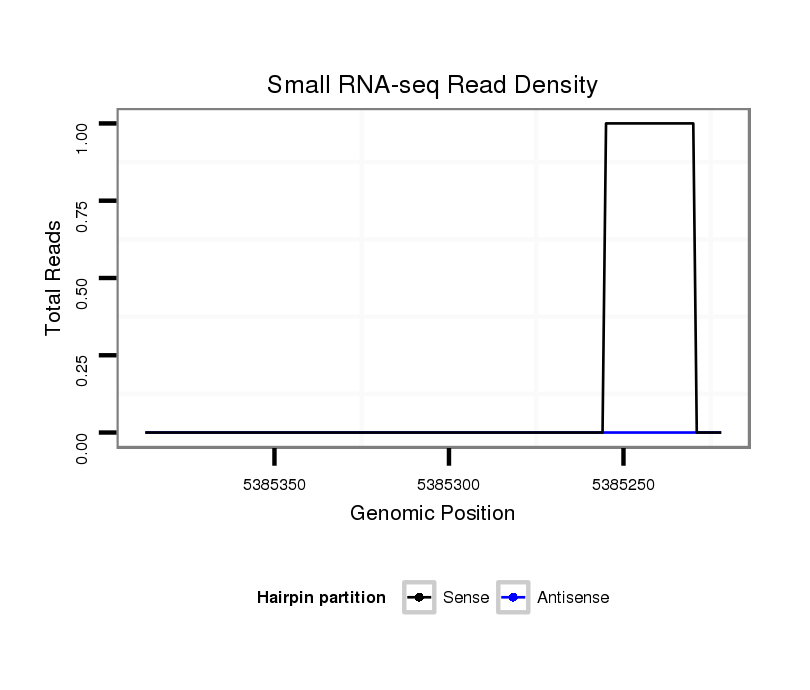
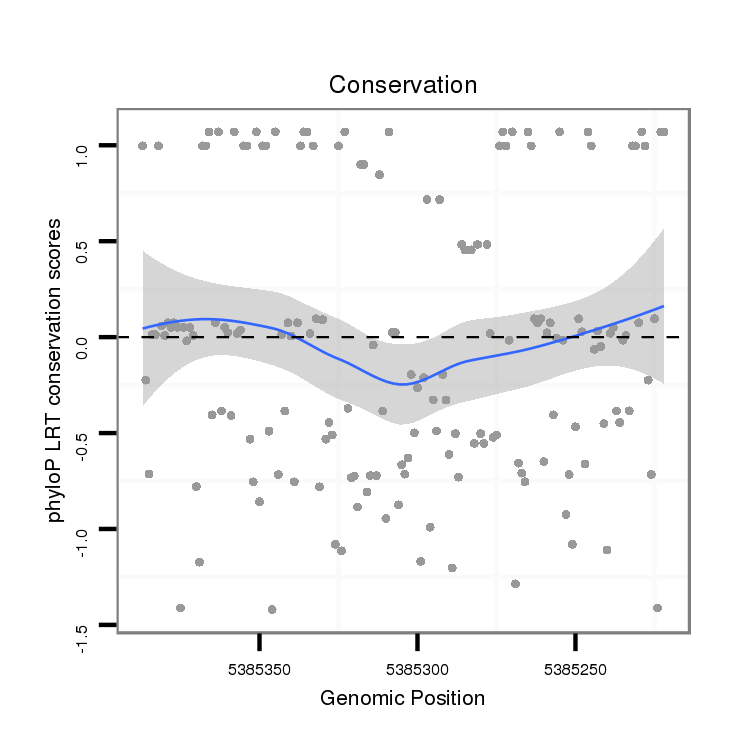
ID:dvi_10569 |
Coordinate:scaffold_12963:5385272-5385337 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_1144:1]; CDS [Dvir\GJ11398-cds]; exon [dvir_GLEANR_1144:2]; CDS [Dvir\GJ11398-cds]; intron [Dvir\GJ11398-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------################################################## CGTCGCCCCACTGTGAGGCGCACCTGAATTACGCCAAGCCGGTGGCGCAGGTATGTGTGAATCGAATTGAAGGAGCCCAAAGGACGAATTTTGCAGCGGTTTCGCATTTTAAACAGCAACGATGTGAAAGGCTGACATTGATGGACAAAAGTCGACGCTGCGACTA **************************************************..(((((.((((((...(((((...((....)).....)))))...))))))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
SRR060666 160_males_carcasses_total |
SRR060680 9xArg_testes_total |
SRR060677 Argx9_ovaries_total |
M047 female body |
V053 head |
SRR060687 9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATGTGTGAATCGAATTGAATGAGC.......................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................GATGGACCAAAGTACACGCT....... | 20 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................TGACATTGATGGACAAAAGTCGACGC........ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................GGAGGGAGCCAAAAGGACGA............................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..TCGCCCCACTGTGACGCC.................................................................................................................................................. | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................CGGGCAAACGTCGACGCTGC..... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................CTGACACTGAGGGACGAAA................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................ACCCGAAGCCGGTGGCCCA..................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GCAGCGGGGTGACACTCCGCGTGGACTTAATGCGGTTCGGCCACCGCGTCCATACACACTTAGCTTAACTTCCTCGGGTTTCCTGCTTAAAACGTCGCCAAAGCGTAAAATTTGTCGTTGCTACACTTTCCGACTGTAACTACCTGTTTTCAGCTGCGACGCTGAT
**************************************************..(((((.((((((...(((((...((....)).....)))))...))))))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060672 9x160_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060681 Argx9_testes_total |
SRR060663 160_0-2h_embryos_total |
GSM1528803 follicle cells |
SRR060666 160_males_carcasses_total |
SRR060687 9_0-2h_embryos_total |
SRR060657 140_testes_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................CCATACACCCTTAGCTTG................................................................................................... | 18 | 2 | 3 | 1.33 | 4 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGCTTCAAATTTGGCGTTGC............................................. | 20 | 3 | 7 | 0.29 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCCATACACCCTTAGCTTG................................................................................................... | 19 | 3 | 15 | 0.27 | 4 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| .....................................................................................................AGCTTCAAATTTGTCGTGGC............................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................GCATAAAAAGTCGCCAAAG............................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGCTTAAAATTTGGCATTGC............................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TTCCTCGGATTTCCT.................................................................................. | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGCTTCAAATTTGTCATTGC............................................. | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................ACCCACTTAGCTAAACGTCC............................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................AAAATTTGAGGTTGCCACA......................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:5385222-5385387 - | dvi_10569 | CGTCGCCCCACTGTGAGGCGCACCTGAATTACGCCAAGCCGGTGGCGCAGGTATGTGTGAATCGAATTGAAG-G--AGC---------CCAAAGGACGAATTTTGCAGCGGTTTCGCATTT--TAAACAGCAACGATGTGAAAGGCTGACATTGATGGACAAAAGTCGACGCTGCGACTA |
| droMoj3 | scaffold_6500:17213915-17214079 + | CGGCGCTTCGCCCCCGAAGGCATCTAGCCTTCGCACATCCGCTAGTGCCGGTAAGTCTTTGGCCA---------------AATGATGTTTAAATGCTGCTCTTATCAGACACGTCGCTTCAAATTTGCAGGATAGCTGTGAGCGAATCGTCTTTCTGGACAGAGAAGAGCGCTGCAAGTA | |
| droGri2 | scaffold_15252:8013896-8014052 - | CATCGCCTCACTATGAAAGGCATCTGAACTTTGCCAAACCATTGGCGAAGGTAAGTGTGAACCAATACAAAT-ATTAAC---------CAATTGCATAAAGCTGGGAACGC-------------TTACAGGAGCTATGTGAAAGAATCTCGCTGCTGGAGTCGAGTGAACGCTGTGATTA | |
| droWil2 | scf2_1100000004884:360221-360373 + | CAAATCCTTATTTTGAACCGCACTTAAATTTCGCTCAGCCTATCTTACCAGTAAGCAAATTTCAAAGACAAATTTCGTCTATTAACATTTAAACATA---------------------------TATCAGGACACCTGCAGCAAGATCGGAACGTTGCCCGCAAAAGAGCGTTGTCGATA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/19/2015 at 04:12 PM