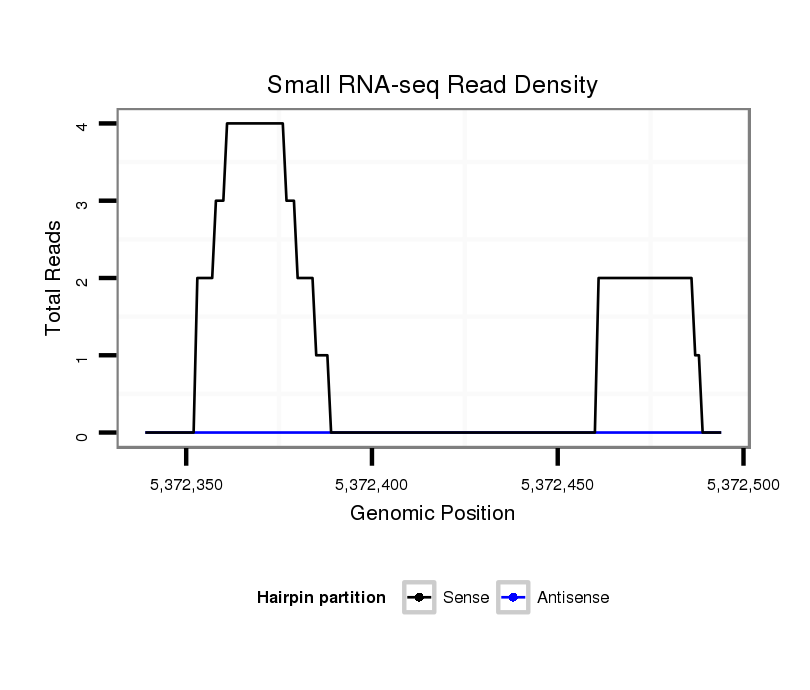
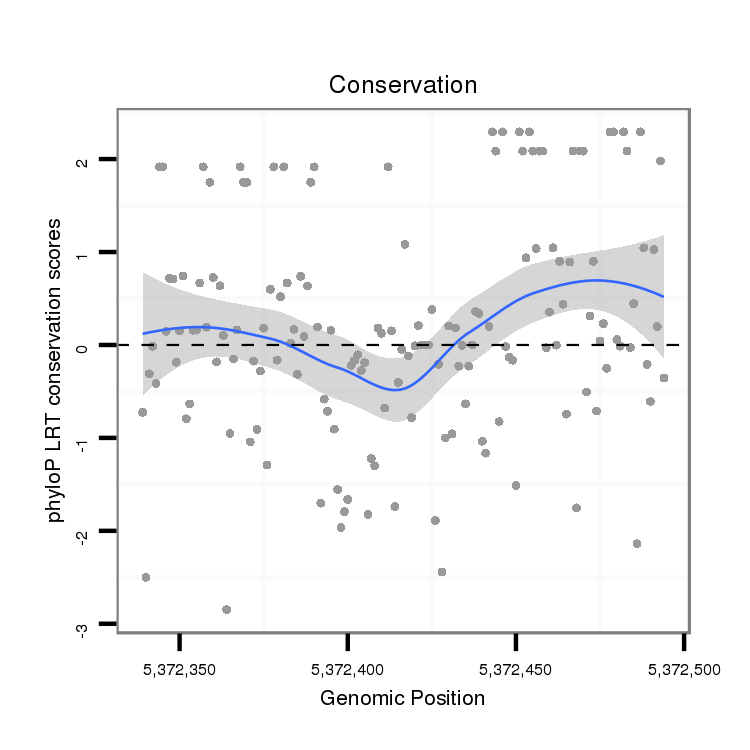
ID:dvi_10566 |
Coordinate:scaffold_12963:5372389-5372444 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_17681:2]; exon [dvir_GLEANR_17681:1]; CDS [Dvir\GJ17153-cds]; CDS [Dvir\GJ17153-cds]; intron [Dvir\GJ17153-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------################################################## GAAAATACTTTGAATATGATGATCAGTTTTGGATTGAGATACAGGCAAAGGTGGATTGCAAAATTGACATATTTATTATAATACATATGTATATTTCTTGTCTCAGTTGGGTTGTTGTGGCCTAGAGGGACCACGAACTTATTTGGACTACTTGCT **************************************************..((((....((((..((((((.(((....))).))))))..))))...))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060670 9_testes_total |
SRR060689 160x9_testes_total |
SRR060655 9x160_testes_total |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
V047 embryo |
SRR060661 160x9_0-2h_embryos_total |
V116 male body |
SRR060660 Argentina_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............TATGATGATCAGTTTTGGATTGAG...................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TAGAGGGACCACGAACTTATTTGGACTA...... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............TATGATGATCAGTTTTGGATTGAGATA................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................TGAGATACAGGCAAAGTTGGGTTGT................................................................................................. | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GGCAAAGTTGGGTTGTTGTGGC................................... | 22 | 3 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TAGAGGGACCACGAACTTATTTGGAC........ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................TGATCAGTTTTGGATTGAGATACAGGC.............................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................TTGAGATACAGGCAAAGTTGGGTT................................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TCAGTTTTGGATTGAGATACAGGCAAAG.......................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................TGAGATGCAGACAAAGGTGGAG.................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................GGTTTTTATGGTCTAGAGGGA.......................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................GGTGAGGTGGCCTAGAGG............................ | 18 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| ...........................................................................................TATTGCTTGTGTCGGTTGGG............................................. | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
CTTTTATGAAACTTATACTACTAGTCAAAACCTAACTCTATGTCCGTTTCCACCTAACGTTTTAACTGTATAAATAATATTATGTATACATATAAAGAACAGAGTCAACCCAACAACACCGGATCTCCCTGGTGCTTGAATAAACCTGATGAACGA
**************************************************..((((....((((..((((((.(((....))).))))))..))))...))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060663 160_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|
| .......................................................................................TCATATAAAGAAGAGAGTCC................................................. | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 |
| ........AAACTTATACTACTACGCAAT............................................................................................................................... | 21 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| ..............................CCTAGGTCTATGTCCGGTTC.......................................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:5372339-5372494 + | dvi_10566 | GAAAATACTTTGAATATGATGATCAGTTTTGGATTGAGATACAGGCAAAGGTGGA--TTGCAAA----------ATTGACAT----A-------TTTATTA-----TAATACATATGTATAT-TTCTT-GTCTCAGTTGGGTTGTTGTGGCCTAGAGGGACCACGAACTTATTTGGACTACTTGCT |
| droMoj3 | scaffold_6500:17223759-17223913 - | AGGATTACTTTGAGAACGATGATCCCAAGTGGTTGGACATGCAGGCGAAGGTATGTGGGCTTCCTTTTTCGAAA-------ATATA---CATCTGGTA-------------------TACGC-TGCTT-TTCTCAGATGGGCTGTTGCGGCTTGAGCGGTCCTCAAACTTACCTGGACTACCTGCG | |
| droGri2 | scaffold_15252:8001789-8001946 + | GCAGATACTTTGAGCATGATGATCAATTTTGGATTGAGATACAGACAAAGGTGCAGAGTGGA----------TAGCAAAA-AT----------ATATCGATTTATCTAATA------CATATATTCCC-TTATCAGTTGCAGTGTTGTGGTTTGGAAAGCCCACGAACTTATCTGATGTACTCGCG | |
| droPer2 | scaffold_5:3903166-3903322 + | TGAAATATATCAATACCAATGACACCTACTGGACGGAACTTGAATTGAAGGTAGTAGTACCTAT----------ATTACTTG----TGAGATCTGGTAA---------TG----AGTGCTCT-CTTCCA-CTGTAGCTGCACTGTTGTGGCTTTGATGGTCCCCGCTGGTACATGCATTACCTGCG | |
| droAna3 | scaffold_12943:2897910-2898067 + | GGAATTACTTTAACTACAACGACACTACTTGGACCCAACTTGAAAAATCGGTACGATATTTAGA----------ATAATTAA----TTAGCTTTTATAA---------TTA---ACTATTTA-ATAATC-TGATAGTTCCATTGTTGTGGATCGGAAGGACCGAGGGATTATTTGGAATTCCTTCA | |
| droKik1 | scf7180000302674:245671-245822 + | CACATTACTGGTGGAGCAACGACACATATTGGTTTAATCTTGAAAAAAAGGTATGATCTCTGGTCTTATAGAGA-------TTAAA----ATCTTATCG---------------AG-------ATGTT-TACTTAGCTGCAATGTTGTGGATTGGGAGGACCCCGAACCTATATGGAGTATCTGCG | |
| droFic1 | scf7180000453973:302276-302340 + | TATA------------------------------------------------------------------------------------------------------------------------TCAT-TTCGCAGCTACAATGCTGTGGATTGGAAGGACCCAGGTCTTATATGGACTACATGCA | |
| droRho1 | scf7180000777421:42363-42519 - | TAAACTACTTCAATTACAATGCCACGAATTGGCCAACGCTTGAAAAGACGGTAAGGTTTCCTCCTTTTTAATTA-------ATACG----ATTTTCTAT----------------AGCATTT-ATCACC-TTTCAGCTTCAATGCTGTGGATTGGAAGGCCCCCGATCCTATATGAGTTATTTGCA | |
| droBia1 | scf7180000302422:3208420-3208573 + | TGAGCTATTTCAATTACAATGATACCTATTGGACTACTCTCGAAAAGACAGTATGACTTCAC----------TAAAA-AGGCT-----------TATACCT-----TTCCT---TATTATCT-TTTATC-TTGCAGATGCTATGCTGTGGACTGGAGGGACCCCGGTCTTATATGGACTACCTACA | |
| droTak1 | scf7180000415858:478717-478871 + | ACAGCTACTTCAATTACAATGAGACGCATTGGCATGATCTTGAAAAGACGGTATCATTTACATA----------GTAGACTC----T-------TATATCT-----TTTTA---ACTGCTCT-TATATC-TTGCAGATGCTCTGCTGTGGACTAGAAGGTCCCCGGTCTTACATGGATTATCTACA | |
| droEug1 | scf7180000409752:369708-369763 - | T----------------------------------------------------------------------------------------------------------------------------------CTCAGATGAGGTGCTGTGGGCTGGAAGGACCGCGGTCCTATATGGACTACATGCT | |
| droYak3 | 2R:907648-907718 + | TTTT-------------------------------------------------------------------------------------------------------AAT-------AGATT-TGCAT-TTCACAGTTGGTATGCTGTGGACTGGAGGGTCCCCGCTCTTATATGGATTATCT--- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/19/2015 at 04:12 PM