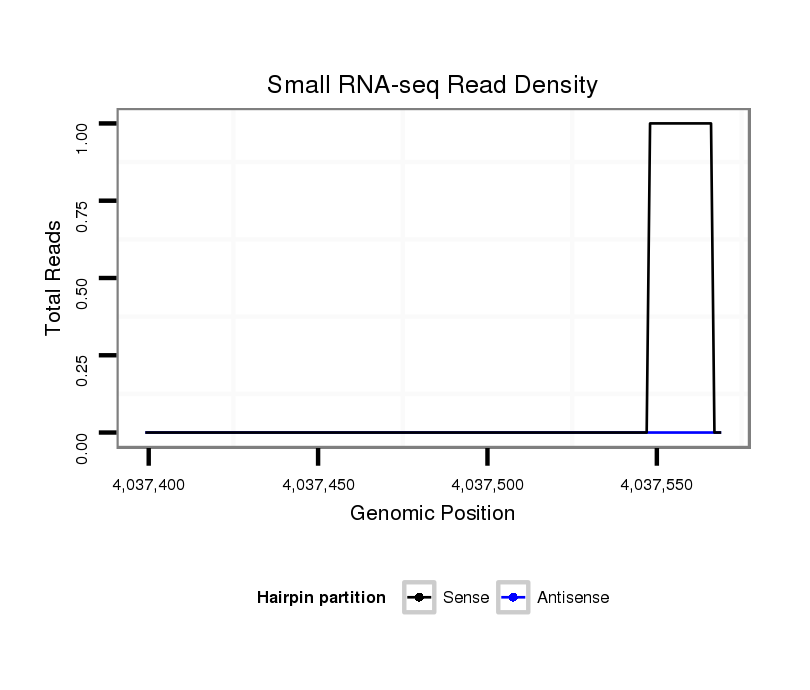
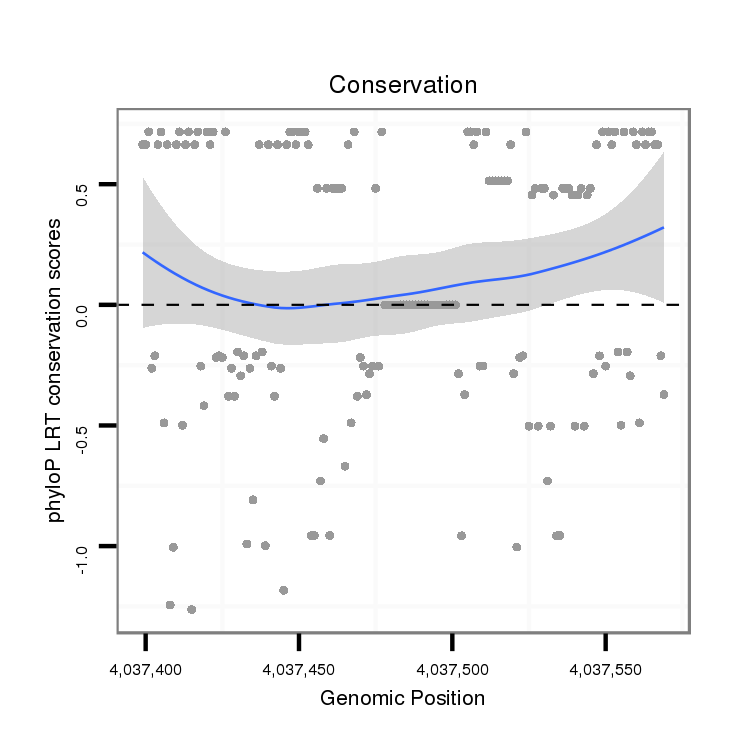
ID:dvi_10439 |
Coordinate:scaffold_12963:4037449-4037519 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ15833-cds]; exon [dvir_GLEANR_1627:4]; exon [dvir_GLEANR_1627:3]; CDS [Dvir\GJ15833-cds]; intron [Dvir\GJ15833-in]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------------################################################## GCTATGAGGCTGAAGAGGATTTGTATGATATCGTTAGAGGCGATCATGATGTAAGGCACATCAAATGGAATTTGATATATTGATTGTTTCTTTGCAAATTGAAATCATGTTTAAATTATAGAACAACGATATTACGCTATCAGAAGATGATTTGACAAGCACGAGTTCCAG **************************************************((((((.((((((((((........))).)))).)))..))))))..........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060662 9x160_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................ATTTGACAAGCACGAGTTC... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................ATATGACAAGGACGAGATCC.. | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................ATTAGACAAGCACGTGTT.... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................AATGATTGTTTTTTTGCAAA......................................................................... | 20 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................TCGCAAACTGAAATCATG.............................................................. | 18 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................AATTGTAGAACAACGGTCT....................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ..............GAGGATTTGTATGAGTTCA.......................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
CGATACTCCGACTTCTCCTAAACATACTATAGCAATCTCCGCTAGTACTACATTCCGTGTAGTTTACCTTAAACTATATAACTAACAAAGAAACGTTTAACTTTAGTACAAATTTAATATCTTGTTGCTATAATGCGATAGTCTTCTACTAAACTGTTCGTGCTCAAGGTC
**************************************************((((((.((((((((((........))).)))).)))..))))))..........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR1106714 embryo_2-4h |
V047 embryo |
M027 male body |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................TCTGCTATACTGTGCGTGCTC...... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .....................ACATACTACAGCAATTTACGCT................................................................................................................................ | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| CGACACTCCGACTTC............................................................................................................................................................ | 15 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................TCTACTTAACTGTTTGTG......... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ..........................CTATGGCAATCTCAGCTG............................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:4037399-4037569 + | dvi_10439 | GCTATGAGGCTGAAGAGGATTTG---TATGATATCGTTAGAGGCGATCATGATGTAAGGCACATCAAATGGAATTTGATATATTGATTGTTTCTTTGCAAATTGAAATCATG-------------------TTTAAATTATAGAACAACGATATTACGCTATCAGAAGATGATTTGAC---------AAGCACGAGTTCCAG |
| droMoj3 | scaffold_6500:7881559-7881719 + | GCTTTGAGGAGGACGACGACATGATGTATAATTTCGTATCAGGAGGTCTGGATGTAAGCGAATTGAAATTGCATTCAGCACA------------------------GCTATGTCTATATT-TTCAACATCTTCCA-------GGCTAATGACATGGCCGTATCGGAGGACGATCTGAC---------CAGCACGAGTTCCAA | |
| droGri2 | scaffold_14906:5097133-5097274 - | GCTACGACGGAGAAGATGATATG---TTCGAGAGTTCGACGGATGAGCACGATGTAAG-----------TGCAGATAGT-TA------------------------GATATGCTGGGACTCCGTGACATCTTTTAAATTATAGGTCGA---------------------CGGTTTGATGACACACAAAAAAACCAGTTCCGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 10:59 PM