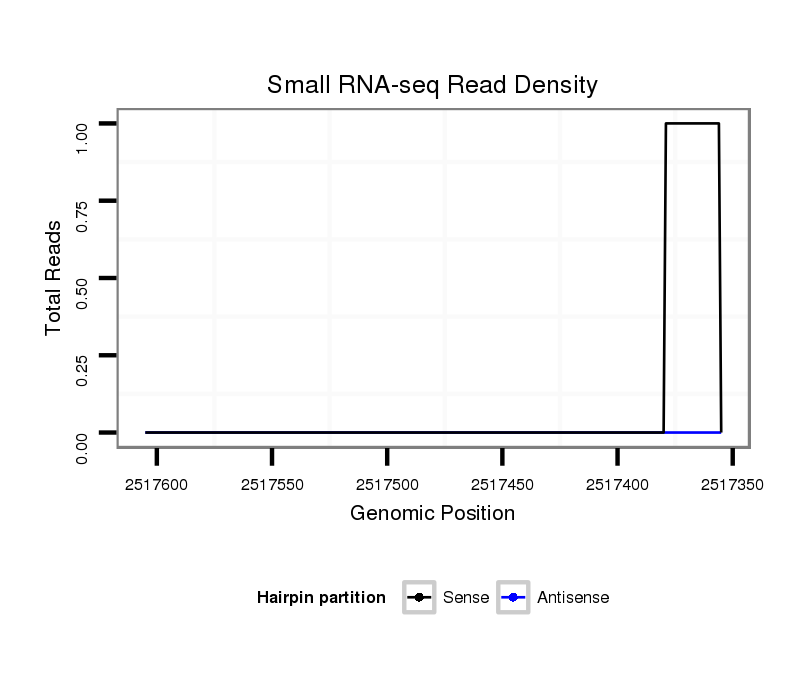
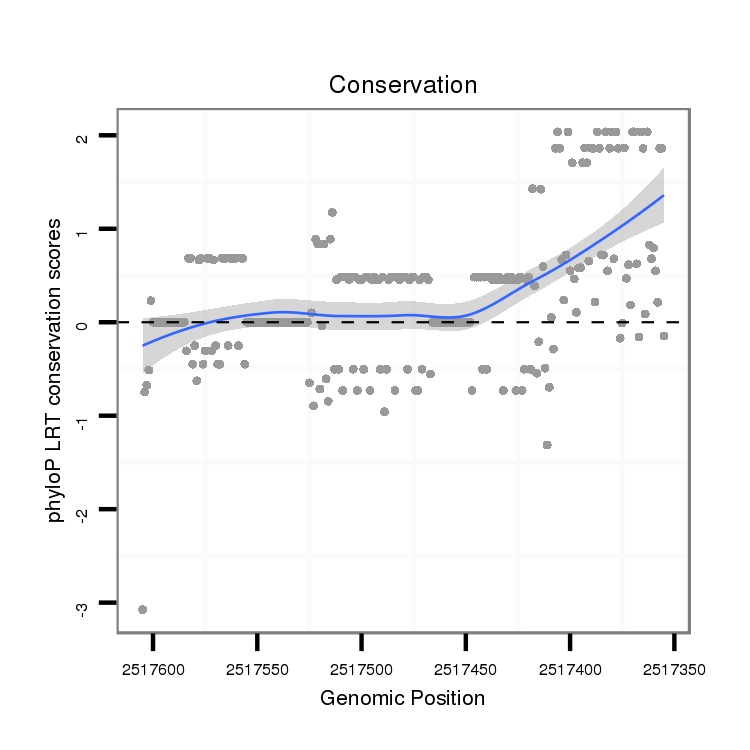
ID:dvi_10197 |
Coordinate:scaffold_12963:2517405-2517555 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ12992-cds]; exon [dvir_GLEANR_1292:3]; intron [Dvir\GJ12992-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACCGACCATTTTCTGGGGTTTATAGCCCTGTAATGCATTTATTAAATTTATTTCGGAACTAAGGCAACTCTGTTCACAGCGATACGTGAGTTTCGTGTAGTGAGCCCTTTCACGCTCCCATATCTAAACAATCCTTAGTCCTTGTTTCATAGCATATTAATTTTTCTTCTCTCGCTCTCGCTACCATTCTCTCCAATGCAGCTGAGCACGGCTCTTCCAGCGATGACACTGTCTCATCGTCGTTTTATGGC **************************************************.....((((..((....))..))))..(((((...(((((...........((((........))))......((((.......))))....((((....))))................)))))..)))))...................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
M061 embryo |
V047 embryo |
SRR060671 9x160_males_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
V053 head |
V116 male body |
M027 male body |
SRR060666 160_males_carcasses_total |
SRR060688 160_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................GTCGGGTAGTGAGGCCTT.............................................................................................................................................. | 18 | 3 | 20 | 6.95 | 139 | 139 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TCGGGTAGTGAGGCCTTG............................................................................................................................................. | 18 | 3 | 20 | 1.50 | 30 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ..................................................................................................................................................................................................................................CACTGTCTCATCGTCGTTTTATGG. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TCGTGTAGTGAGGGCTTT............................................................................................................................................. | 18 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................GGGTCGGGTAGTGAGCCC................................................................................................................................................ | 18 | 3 | 15 | 0.27 | 4 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GTCGGGTAGTGAGGCCTTT............................................................................................................................................. | 19 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................AGTTTCGGGTAGTGAGGGC................................................................................................................................................ | 19 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................GCCCCCTCTACCATTCTCTCC......................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................AGGGTCGTGTAGTGAGGCC................................................................................................................................................ | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CTTCCACCGTTGAAACTGT................... | 19 | 3 | 18 | 0.11 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................TCCCATATCTCAACAGGCC..................................................................................................................... | 19 | 3 | 19 | 0.11 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................TCTTCCACCGATGTCACTT.................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................GGGTCGTGTAGTGATCCC................................................................................................................................................ | 18 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TGAGGGTCGGGTAGTGAGC.................................................................................................................................................. | 19 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TCACAGCCATACGTGAAGT............................................................................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TACTGCGCCCTTTCAAGCT....................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
TGGCTGGTAAAAGACCCCAAATATCGGGACATTACGTAAATAATTTAAATAAAGCCTTGATTCCGTTGAGACAAGTGTCGCTATGCACTCAAAGCACATCACTCGGGAAAGTGCGAGGGTATAGATTTGTTAGGAATCAGGAACAAAGTATCGTATAATTAAAAAGAAGAGAGCGAGAGCGATGGTAAGAGAGGTTACGTCGACTCGTGCCGAGAAGGTCGCTACTGTGACAGAGTAGCAGCAAAATACCG
**************************************************.....((((..((....))..))))..(((((...(((((...........((((........))))......((((.......))))....((((....))))................)))))..)))))...................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060687 9_0-2h_embryos_total |
V116 male body |
M061 embryo |
M047 female body |
SRR060668 160x9_males_carcasses_total |
SRR060689 160x9_testes_total |
SRR060658 140_ovaries_total |
M027 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................CGGAGAAGGTCGCTATTCTG...................... | 20 | 3 | 6 | 0.67 | 4 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................GGAATAGATAAGTTAGGAATC................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................AGCACATCACTCGAGTAATT........................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................AGAAGGTCGCTACCATGATA................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................AAGAGGGGTTACGTCGTC............................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................AGAAGGTCGGTATTCTGAC.................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................AAAGTCCGAGAGTATAGCT............................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:2517355-2517605 - | dvi_10197 | ACCGACCATTTTCTGGGGTTTATAGCCCTGTAATGCATTTATTAAATTTATTTCGGAACTAAGGCAACTCTGTTCACAGCGATACGTGAGTTTCGTGTAGTGAGCCCTTTCACGCTCCCATATCTAAACAATCCTT---------------------AGTCCTTGTTTCATAGCATATTAATTTTT-CTTCTCTCGCTCTCGCTACCATTCTCTCCAATGCAGCTGAGCACGGCTCTTCCAGCGATGACACTGTCTCATCGTCGTTTTATGGC |
| droMoj3 | scaffold_6500:12691343-12691520 - | ATAGA---------------------------------------------------------------------------TACACTTGCTTTCCATTTAGTAATCCTTTGCACGTTGTCATCTCTAAGCAAGACCTTTTGGATATTTGTATTAATCCAGA-------------------CATTTCTCTTTCTCTCTCTCTCTCTCTCACTCAC-TATCTTCAGCTGAGCACGGCTCTTCCAGCGATGACACGGTCTCATCGTCGTTTTATGGC | |
| droGri2 | scaffold_15252:2126497-2126561 - | TT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCTTTGTTGCAGCTGAGCACGGCTCTTCCAGCGATGACACCGTCTCATCGTCGTTTTATGGA | |
| droWil2 | scf2_1100000004521:6782025-6782124 - | AATTT----------------TTATTGCTTAAAAGTCGTTACTAAACTTC------------------------------C----------T---------------------------------------------------------------------------------------------------------------------TCCTTTCCTTACAGCTGAGCACGGCTCTTCCAGCGATGACACCGTCTCATCGTCGTTCTATGGC | |
| dp5 | 4_group5:3601-3661 - | TC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTCCACAGCAGAGCCCGGCTCTTCCAGCGATGACACCGTCTCATCGTCGTTCTACGGC | |
| droPer2 | scaffold_5:4503268-4503348 - | CCTCT---------------------------------------------------------------------------TAGACTCGAATT---------------------------------------------------------------------------------------------------------------------TCCATTCTCCACAGCAGAGCCCGGCTCTTCCAGCGATGACACCGTCTCATCGTCGTTCTACGGC | |
| droAna3 | scaffold_13258:42049-42102 + | G---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGCAATCCCAGCTCATCTAGTGATGACACCATTGAATCTTCATTCTATGGA | |
| droBip1 | scf7180000396329:10225-10282 - | CA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCGCAGCCAATCCTAGCTCATCTAGTGATGACACCGTTGCATCATCTTTCGATGGA | |
| droKik1 | scf7180000302644:30663-30733 + | CTTGT---------------------------------------------------------------------------TG-------------------------------------------------------------------------------------------------------------------------------TTTTTTTCTTACAGCTGAGCCTGACTCTTCTAGCGACGACACACTTTCATCGTCTTTCTGCGGC | |
| droFic1 | scf7180000453851:273828-273875 + | TTC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTCACAGCTGA-----------CCAGCAACGAAACCC---TATGCTCTTCCTGTGGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
Generated: 05/19/2015 at 04:04 PM