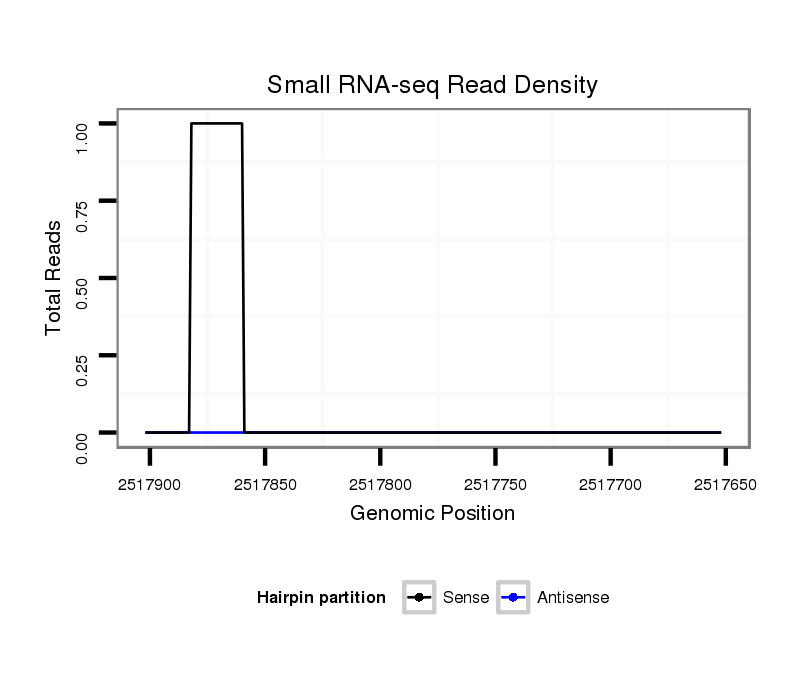
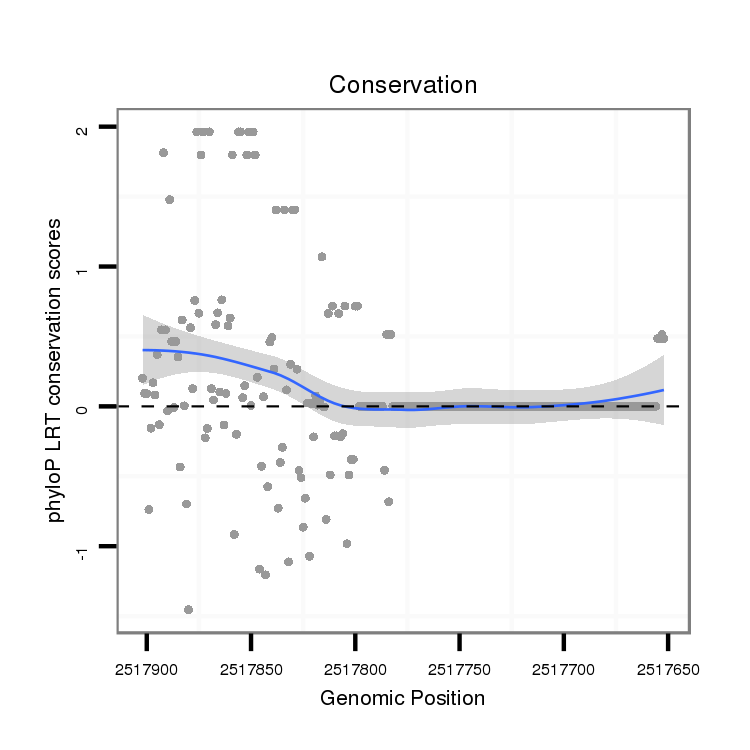
ID:dvi_10196 |
Coordinate:scaffold_12963:2517702-2517852 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_1292:2]; CDS [Dvir\GJ12992-cds]; intron [Dvir\GJ12992-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AGCCTGCGGCACACAGACCTGTGGCAAGGAACTGGGCGTCAGCGCCAAAGGTGAGTTATGCAAAATATTCGAATTATTGAGATGGGAGCGAAAAGTGAGAAAAAGTGAATACCCAAGAGACAAAAAAAAAAATATTAGTATATATAACATTTTTGCACAATTTAATGAGGATTCTTGTCAGTAACAAGCTAGGATTGCCAAGAACCTTTTTGCACAGAAAACTTTATCTATGTATTAATCAAATAATCGAC **************************************************..........................((..(((...(((......(((.((.....((((.((((..(((.......((((((.(((.......))).))))))......)))..)).)))))))).))).......)))...)))...))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060667 160_females_carcasses_total |
SRR060673 9_ovaries_total |
SRR1106720 embryo_8-10h |
V047 embryo |
V053 head |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................AAGAACCTTTTTGCACAGAAAATTTAATCTATGTATTA.............. | 38 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................GTGGCAAGGAACTGGGCGTCAGC................................................................................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AGTGAATGCCCAAGAGACCA................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................GGCGTTAGCGTCAAGGGTGA..................................................................................................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................CGGAACTGGGCTTCACCGC.............................................................................................................................................................................................................. | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................CAAGAACCTTTTGGAACA................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............AAGTCCTGTGGCAAGGAGCT.......................................................................................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................GTGTGGCGAGGAACTCGG....................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................CAACGTGACTACCCAAGA..................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TCGGACGCCGTGTGTCTGGACACCGTTCCTTGACCCGCAGTCGCGGTTTCCACTCAATACGTTTTATAAGCTTAATAACTCTACCCTCGCTTTTCACTCTTTTTCACTTATGGGTTCTCTGTTTTTTTTTTTATAATCATATATATTGTAAAAACGTGTTAAATTACTCCTAAGAACAGTCATTGTTCGATCCTAACGGTTCTTGGAAAAACGTGTCTTTTGAAATAGATACATAATTAGTTTATTAGCTG
**************************************************..........................((..(((...(((......(((.((.....((((.((((..(((.......((((((.(((.......))).))))))......)))..)).)))))))).))).......)))...)))...))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060688 160_ovaries_total |
SRR060666 160_males_carcasses_total |
V047 embryo |
M047 female body |
SRR060662 9x160_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................AAATTACTCCCTAGATCAGT....................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................ACAAGTCTATCCTCGCTTT.............................................................................................................................................................. | 19 | 3 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ......................................................................................TGGCTTTTCGCTCGTTTTCAC................................................................................................................................................ | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................ATGGGATCTCTTTTTTTCTTTT........................................................................................................................ | 22 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................TCCCGCAGTCCAGGTTTC......................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................CTTATTGGTTCTCTGTTATC............................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:2517652-2517902 - | dvi_10196 | AGCCTGCGGCACACAGACCTGTGGCAAGGAACTG---GGCGTCAGCGCCAAAGGTGAGTTATGCAAAAT-ATTCGAA-TTATTGA--------------GATGGGAGCGAAAAGTGAGAAAAAGTGAATACCCAAGA-GACAAAAAAAAAAATATTAGTATATATAACATTTTTGCACAATTTAATGAGGATTCTTGTCAGTAACAAGCTAGGATTGCCAAGAACCTTTTTGCACAGAAAACTTTATCTATGTATTAATCAAATAATCGAC--------------------------------------- |
| droMoj3 | scaffold_6500:12691929-12692030 - | AGCCAGCGCCACACAAACTTGCAGCAAGGAATTG---GGCGTCAGCGCCAAAGGTGAGTGAAAAAGAAA--CTCTAAGTT--CTA--------------TATGAGATGGCAAAGTGAACAAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15252:2127147-2127314 - | AGCCAGCGGCACCCAGACCTGTGGCAAGGAGTTG---GGCATCAGCGTCAAAGGTAAGAAGAAACAAAGTTCTCCAATTTGAAGTGTTTAAAGTTAGCAAGAGAAATGGCAGGGAAATCCCAA------------AAATA-------------------------------------------------------------------------------------------------------------------------------CGACTCAGAAATAATTATTGTAATTATTTTCTTGATTACTTTA | |
| droWil2 | scf2_1100000004521:6783166-6783249 - | AGCGAATGGCACACAGACCTGTGGCAAGGAATTG---GGCGTAAGCGCTAAGAGTAAGTACAAAAAGAA--TTG-AAATA--TTA--------------TA-AAGAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | 4_group5:3711-3769 - | AGCAAGCGGCACACAGACGTGCAGCAAAGAGTTGCAAGGCGTAGGCGGAAAGGGTGAGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_5:4503378-4503436 - | AGCAAGCGGCACACAGACGTGCGGCAAAGAGTTGCAAGGCGTAGGCGGAAAGGGTGAGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13258:13186-13255 + | AACAAGCTTTAT-------TTTCCTAAGGAGATA---AGTCTAAGCGTAAACGGTAAGTTGACAAAAAAAATTCCGAATT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302644:28800-28845 + | GCACGA--------------AAAGAGAGGAATTT---TCCGTGGGAGCCAAGGGTAAGATATG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000772993:4697-4759 + | TGGTTGTTTCACGCGAGAGATCAGCAAGGAGCTG---AGCGCGGCCGACAAGTGTAAGTCAATCAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/19/2015 at 04:05 PM