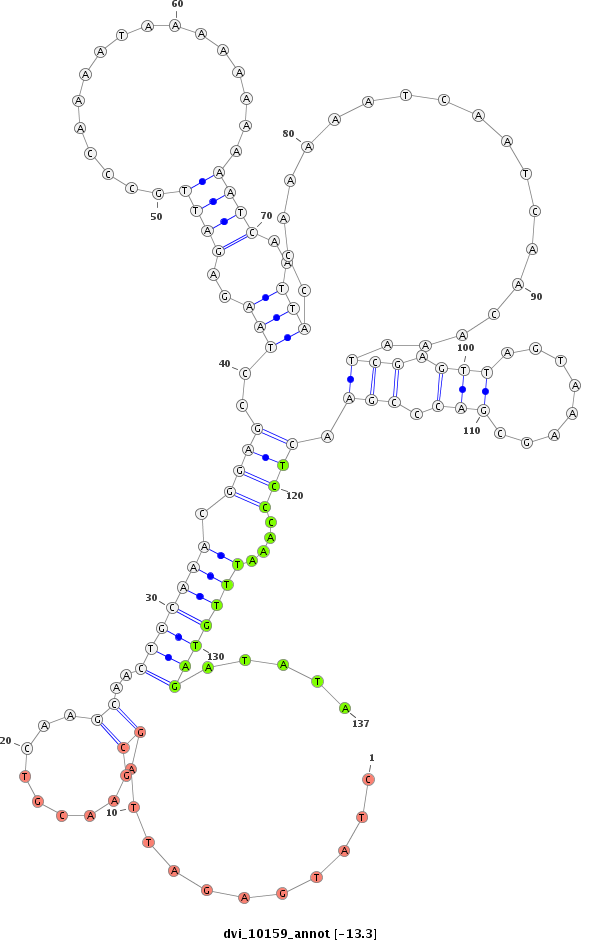
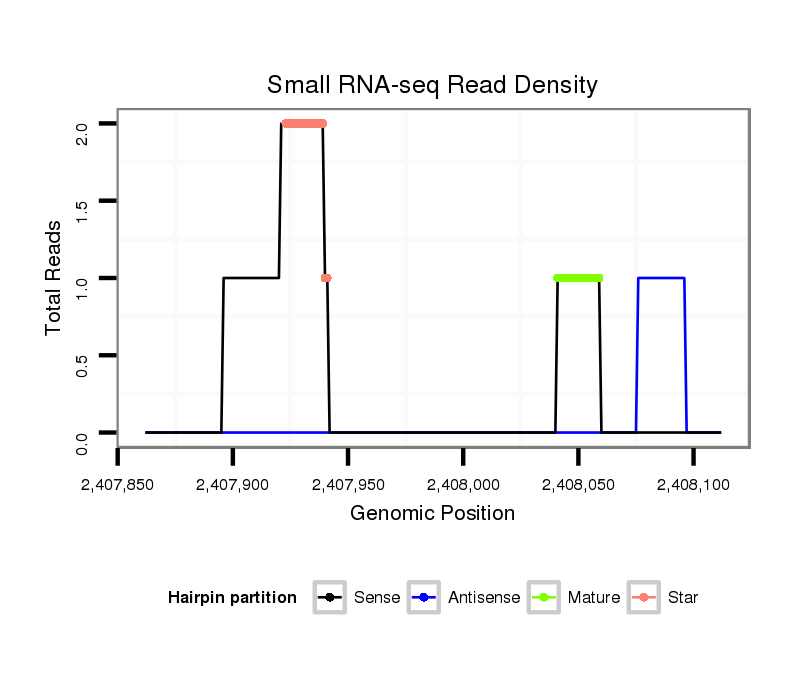
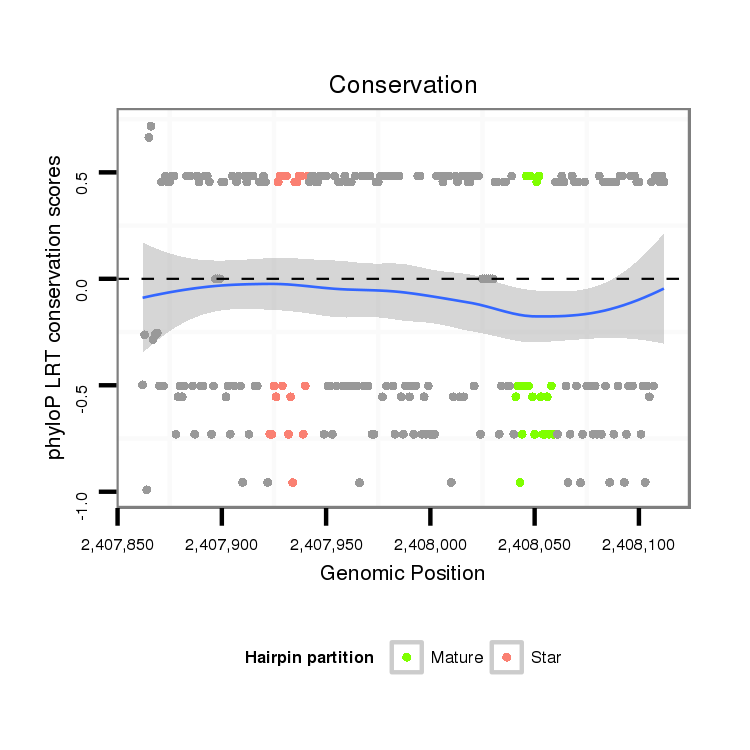
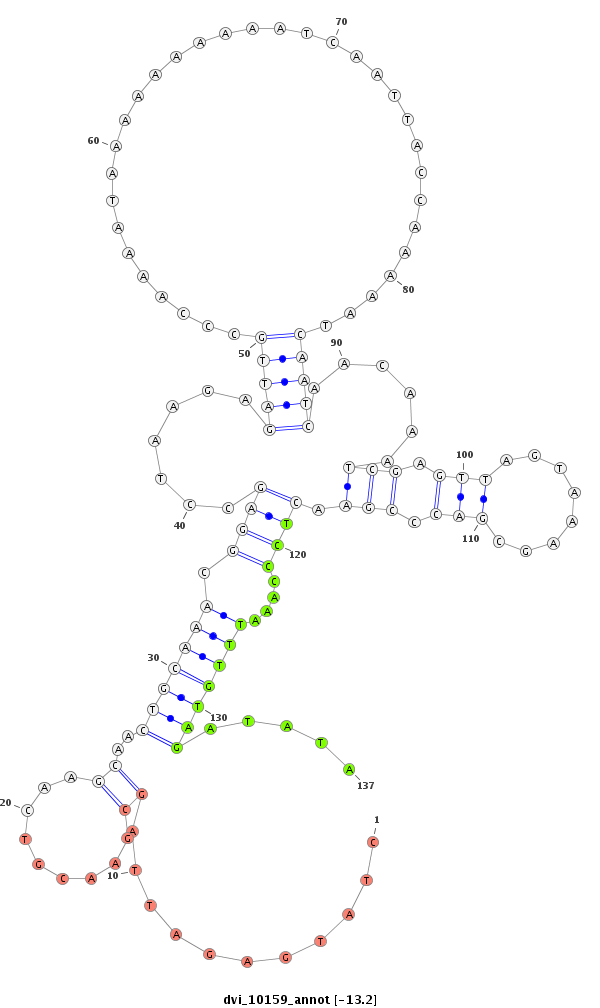
ID:dvi_10159 |
Coordinate:scaffold_12963:2407912-2408062 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -13.2 | -12.9 | -12.9 |
 |
 |
 |
exon [dvir_GLEANR_1528:2]; CDS [Dvir\GJ14968-cds]; intron [Dvir\GJ14968-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTTCAATTCCTACCAACTTTAATTTCACCTAAGCATAACGTCAAGCAACAGCAAACGGAGCCTATGAGATTAGCGAACGTCAAGCAACTGCAAACGGAGCCTAAGAGATTGCCCAAAATAAAAAAAAAATCAATTACCAAAAATCAATCAACAAATCGAGTTAGTAAAGCGACCCGAACTCCCAAATTTGTAGATATACAGAGAGCAGAGGCCGATATCTTCTCCCGCCTACAAATACCAACATGTATCTG *************************************************************...........((.........))..(((((((.((((..(((..((((.................))))..)))...................(((.(((........))).))).))))....))))))).....***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060655 9x160_testes_total |
SRR060657 140_testes_total |
V053 head |
M047 female body |
SRR060678 9x140_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................ATAACGTCAAGCAACAGCAAACGGAGC.............................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................TCCCAAATTTGTAGATATA..................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................CTATGAGATTAGCGAACGT........................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................GCCTATGAGATTAGCGAAC............................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................ATCTGCTCTCGCCTACAAAG............... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................GCGAACGGCAAGCAAGTG................................................................................................................................................................. | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................ACATAGCCTATGAGGTTAG.................................................................................................................................................................................. | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AAAGTTAAGGATGGTTGAAATTAAAGTGGATTCGTATTGCAGTTCGTTGTCGTTTGCCTCGGATACTCTAATCGCTTGCAGTTCGTTGACGTTTGCCTCGGATTCTCTAACGGGTTTTATTTTTTTTTTAGTTAATGGTTTTTAGTTAGTTGTTTAGCTCAATCATTTCGCTGGGCTTGAGGGTTTAAACATCTATATGTCTCTCGTCTCCGGCTATAGAAGAGGGCGGATGTTTATGGTTGTACATAGAC
*****************************************************...........((.........))..(((((((.((((..(((..((((.................))))..)))...................(((.(((........))).))).))))....))))))).....************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060684 140x9_0-2h_embryos_total |
V116 male body |
SRR060655 9x160_testes_total |
V053 head |
M047 female body |
SRR060659 Argentina_testes_total |
SRR060666 160_males_carcasses_total |
SRR060687 9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................TTAGTTAAAGGTTTTTAGT......................................................................................................... | 19 | 1 | 4 | 1.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TATAGAAGAGGGCGGATGTTT................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............TGAAACTAAAGAGGATTC.......................................................................................................................................................................................................................... | 18 | 2 | 8 | 0.25 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GGTTTTGTTTTTTTTTTAGGTAA.................................................................................................................... | 23 | 2 | 8 | 0.25 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................GGATTAGTAGTGCGGTTCGT............................................................................................................................................................................................................ | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................CTGGGCTTGGTGGTTTAAA.............................................................. | 19 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............TGAAACTAAAGTGCATTC.......................................................................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................CGGCGGGTGTGTATGGTTGTA....... | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........ATGGCTGAAATTCATGTGG.............................................................................................................................................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............TGAAACTAAAGAGGATTCC......................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:2407862-2408112 + | dvi_10159 | TTTCAATTCCTACCAACTTTAATTTCACCTAAGCATAACGTCAAGCAACAGCAAACGGAGCCTATGAGATTAGCGAACGTCAAGCAACTGCAAACGGA-------------GCCTAAGAGATTGCCCAAAATAAAAAAAAAATCAATTACCAAAAATCAATCAACAAATCGAGTTAGTAAAGCGACCCGAACTCCCAAATTTGTAGATATACAGAGAGCAGAGGCCGATATCTTCTCCCGCCTACAAATACCAACATGTATCTG |
| droMoj3 | scaffold_6500:12565286-12565540 + | GAACAGACTCCACCAAAACAGATTCAACTCAAGAG---CGATCAACAGGAGAAAGTGGAGGAGGAGAAATGTCCGAAAATCAAGCAAATATCAGCGAATTCAGTGAATCCAGTCCAGCAAACTTACCATAGTAGCAATCGGTCAGATGTATCCCAATCAATGTACTATTCGAATTC------CGCTCCAGAAATGAGAGTAGGTTTCACCCCCGAGGCAAGGGCACAGCCCATGCCCGCTCTTAGCGACACCCGGGAGCATCTG | |
| droGri2 | scaffold_15126:2607157-2607164 - | TTGCAGTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 03:59 PM