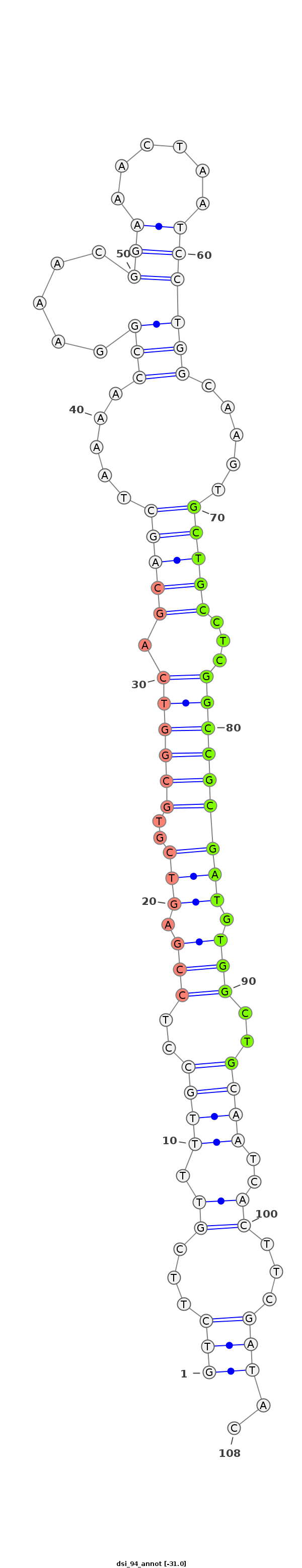
| droSim3 |
2L:913978-914156 + |
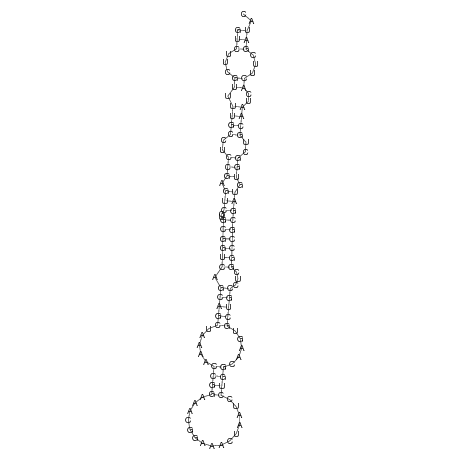
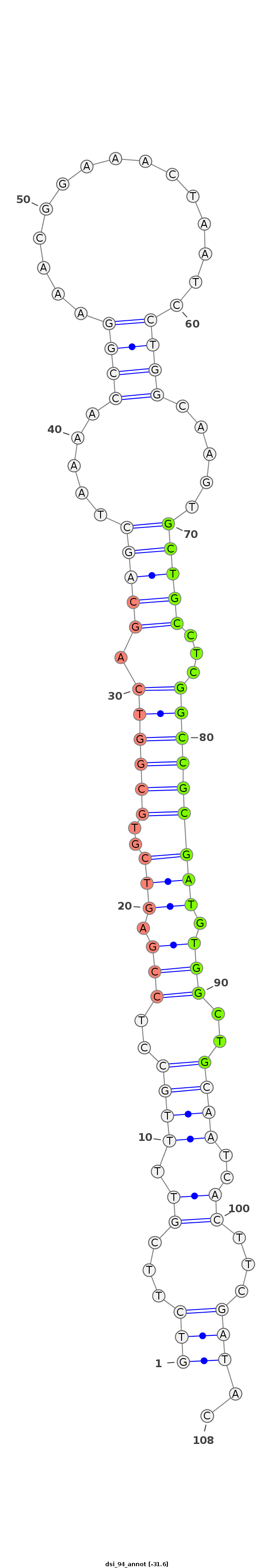
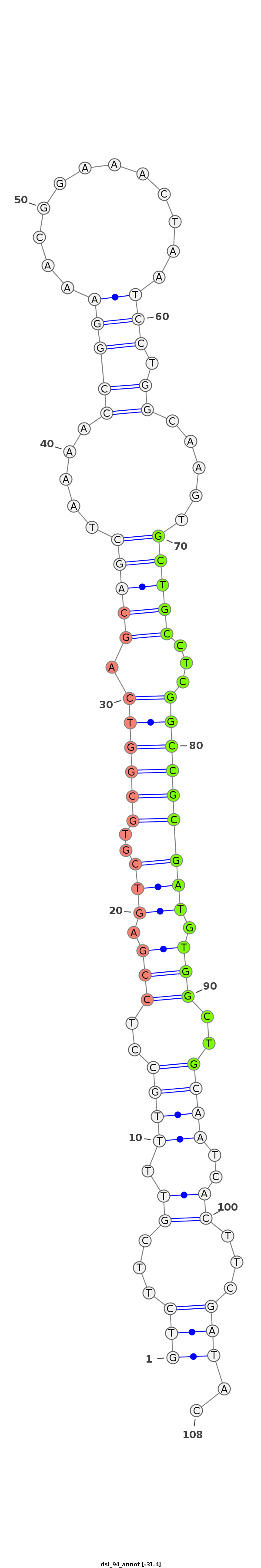
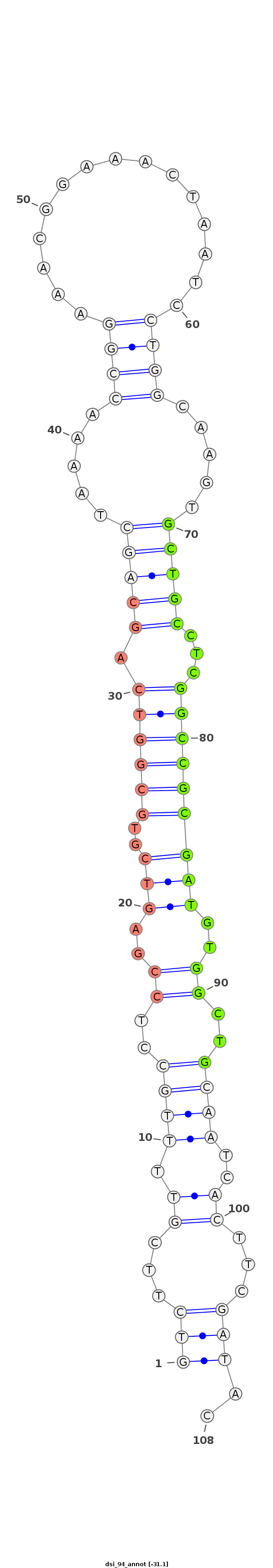
dsi_94 |
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGCTAA-A-ACCGGA-AACGGA--------------AAC-----TAA--TCCTG---------------------GCAAG---------TGCTGCCTCG------GCCGCGATGTG----------------------------------------------------------GCTG----CAA------TCACTTCGATACC-GATTCAGAATCCGCTTC-GATTAGAATTCAGA---------------ATA---------------T-------- |
| droSec2 |
scaffold_14:894220-894398 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGCTAA-A-ACCGGA-AACGGA--------------AAC-----TAA--TCCTG---------------------GCAAG---------TGCTGCCTCG------GCCGCGATGTG----------------------------------------------------------GCTG----CAA------TCACTTCGATACC-GATTCAGAATCCGCTTC-GATTAGAATTCAGA---------------ATA---------------T-------- |
| dm3 |
chr2L:922761-922939 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGCTAA-A-ACCGGA-AACGGA--------------AAC-----TAA--TCCTGGCC---------------------AG---------TGCTGCCTCG------GCCGCGATGTG----------------------------------------------------------GCTG----CAA------TCACTTCGATAGC-GATTCAGAATCCGCTTC-GATTAGAATTCAGA---------------ATA---------------T-------- |
| droEre2 |
scaffold_4929:985342-985533 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGCTAA-A-ACCGGA-AACGGA--------------AAC-----AAG--TCCTGGCCAGCT--AAAG------TGGCAAG---------TGCTGCCTCG------GCCGCGATGTG----------------------------------------------------------GCTG----CAA------TCCTTTCGACGCC-GATTCAGACTGCGCTTC-GATTAGAATTGAGA---------------ATA---------------T-------- |
| droYak3 |
2L:910182-910401 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGCTAA-A-ACCGGA-AACGGAA--ACGGCAACGCCAAC-----AAA--TCCTGGCCAGCC--AAAG------TGGCAAG---------TGCTGCCTCGGTACCAGCCGCGATGTG----------------------------------------------------------GCTG----CAA------TCATTTCGATGCC-GATTCAAACTGCGCCTC-GATTAGAATCCAGA---------------ATAT-------------ATATGGCTGA |
| droEug1 |
scf7180000409554:1652206-1652411 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGCTGG-A-ACCGGA-AACCGGAAATCGGAAACGGAAAC-----TAA--ACCTGTCCAGAT--TGAG------TGGCAAG---------TGCTGC-TCG------GCCGCGATGTG----------------------------------------------------------GCTG----CTA------TCAATTCGGTCCC-GATTCAGATTCCGATTC-GAGTAGATT----------ACTAA----AGTA---------------T-------- |
| droBia1 |
scf7180000302261:2876178-2876390 - |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGTTAA-A-ACCGGA-AACGGAA---------------C-----TAG--ATCTGGCCAAAAT-AGAG------TGGCAAG---------TGCTGC-TCG------GCCGCGATGTG----------------------------------------------------------GCTG----CAATCATCATCATTTCGCCTGC-GATTCAGATACCGATTC-GACTAGAAT----------ACCAA----GATATAACCACTAAATACAG-------- |
| droTak1 |
scf7180000414027:55946-56147 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGCTAA-AAACCGGA-AACGGA--------------AAC-----A-GAAAAC---------------------TGCCAAG--------TGGCTGCCTCG------GCCGCGATGTG----------------------------------------------------------ACTG----CAGTTATTATCATTTCGATACC-GATTCAGATACCGATTC-GATTAGAAT----------ACTATAA--AGTAT----ACAAAATAAAT-------- |
| droEle1 |
scf7180000490458:351461-351666 - |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCTACTAA-A-ACCGGA-AACTGA--------------G-------------TCTGGCCAAAATAAAAAAAAGAATGGCAAT---------TGCTGCCTCG------GCCGCGATGTG----------------------------------------------------------GCAC----CAA------TCATTTCGATTCT-GATTCCGATTACGATTC-CGTT-----TCAGTAGAATACCAGAAAAGATAT-------------AT-------- |
| droRho1 |
scf7180000769144:17225-17423 - |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAT-TTCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCTACCAA-GAACCGGA-AAC-------------------------TAA--ACCTGGCCAAAA--AGAA------TGGCCAA---------TGCTGCCTCG------GCCGCGATGTG----------------------------------------------------------GCTC----CAA------TCATTTCGATTCT-GATTCTGAGTCCGGTTCCGATTTGATTCGAGTAGAATACGAT----AGTA---------------T-------- |
| droFic1 |
scf7180000453904:1194588-1194762 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAC-TCCAGTCTTCGTTTTGCCTCCGAGTCGTGCGGTCAGCAGCTGA-A-CCCCGA-CAC-------------------------TAG--ACCTGGCCAACAA-AATA------AGGCAAG---------AGCTGCCTCG------GCCGCGATGTG----------------------------------------------------------GCTC----AAG------TCACTCTAAC-------------TCCGATTC-GAGCAGAGC----------TCCAG-----AGA---------------C-------- |
| droKik1 |
scf7180000301725:57225-57437 - |
|
TGGTCCTGGTCTCGTATATAACAACGAGTGCGCCAACTGGCCGAC-GCCAGTCTTCGTTTCGCCTCCGAGTCGTGCGGTCAGCAGCTGG-A-ACTGGGTAACAGG--------------GATTTTTGTCA--ACCTGGCCGAAGCA-----------GGCA--------------------G------GTCGCGATGCGACGCCTCAGACTGCACTCCGCCTCGCTCTAGTACATATATATATATATCTCCCACTCGATAGC-------------------------------------------------------GGG---------------ATA---------------T-------- |
| droAna3 |
scaffold_12916:2645190-2645391 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGGCTGAC-TCCAGTCTTCGTTTTGCCTTCGAGTCGTGCGATCAGCAGCGGT-A-ACCGGA-AACTGAA---------------C------------TAGGCCGGAACTGAAGTGGCAATCGCAAGT------GGCGCTGCCTCG------GTCGCGATGCG----------------------------------------------------------GCTA----TAG------AAACCCCAACTTTAAATTCAAA------ATC-GAGTAGAGTCG--A---------------AT--AATCGCCAAA-AGAA-------- |
| droBip1 |
scf7180000396737:2115681-2115828 - |
|
TA------------TATATAACAACGAGTGCGCCAACTGGCTGGCGTCCAGTCTTCGTTTTGCCTTCGAGTCGTGCGATCAGCAGCGGG-A-ACCGGA-AACTGAA---------------C------------TGTGCCGGAACTGAAGTGGCAATCGCAAGT------GGCGCTGCCTCG------GCCGCGATGCC----------------------------------------------------------GCT------------------------------------------------------------------------------------------------------ |
| dp5 |
4_group2:415636-415757 - |
|
TA--------------TATAACAACGAGTGCGCCAACTGAGCGCT-TTCAGTCTTCGTTTTGCCTCTGAATCGTGCGGTCAGCAGCTGG-A--------AACTGAA---------------C------------T-----------------------GCAA-CCCACAGCGCGCTGCCTCG------GCCGCGATGCG----------------------------------------------------------ACCG----------------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_8:3473007-3473128 + |
|
TA--------------TATAACAACGAGTGCGCCAACTGAGCGCT-TTCAGTCTTCGTTTTGCCTCTGAATCGTGCGGTCAGCAGCTGG-A--------AACTGAA---------------C------------T-----------------------GCAA-CCCACAGCGCGCTGCCTCG------GCCGCGATGCG----------------------------------------------------------ACCG----------------------------------------------------------------------------------------------------- |
| droWil2 |
scf2_1100000004585:2154193-2154263 - |
|
TA--------------TATAACAACGAGTGCGCCAACTATGTCAA-TTCAGTCTTCGTTTTGCCTCTGAATCGTGCGGTTAAAAGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droVir3 |
scaffold_12963:19490754-19490884 + |
|
TA--------------TAAAATAACGAGTGCGCCAACTGGCCAAA-TTCAGTCTTCGTTTTGCCACTGAATCGTGCGGTCAGCAATTTGTT--------------------------------------------------------AAGCTGCGACTGCGAATCCA---CGCGTTGCGTCG------GCCGCGACGCG----------------------------------------------------------ATTGGCTGAAA------T--------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6500:23718611-23718740 - |
|
TA--------------TAAAATAACGAGTGCGCCAACTGGCGGAT-TTCAGTCTTCGTTTTGCCACTGAATCGTGCGGTCAACAATTTGTT------------------------------------------------CACA---------GCGACTGCGAATTCA---CGCGTTGCGTCG------GCCGCGACGCG----------------------------------------------------------ATTGGCTGAAA------T--------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15252:7342250-7342377 + |
|
TA--------------TTTAATAACGAGTGCGCCAACTGGCTGAA-TTCAGTCTTCGTTTTGCCACTGAATCGTGCGGTCAGCAATTGA-TAACTGGA-----------------------C-----------------------------------TGCAAATCCA---CGCGCTGCGTTC------GCCGCGCCGCG----------------------------------------------------------ATTGGCTGAAA------T--------------------------------------------------------------------------------------- |