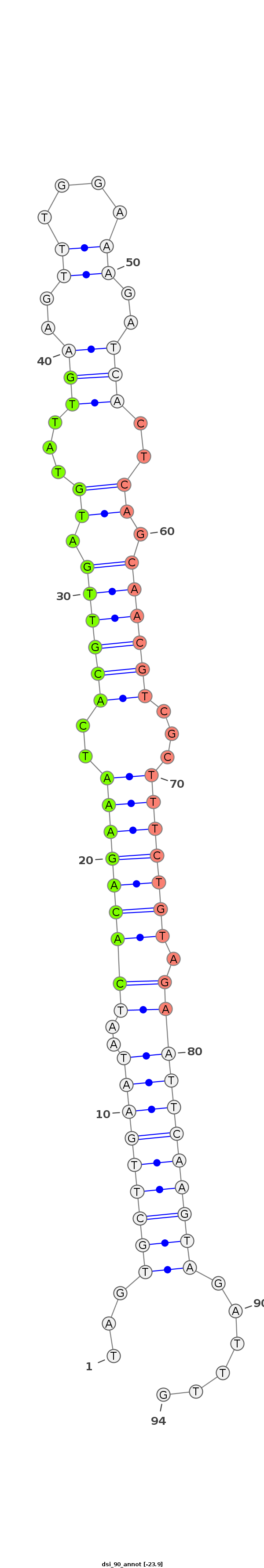
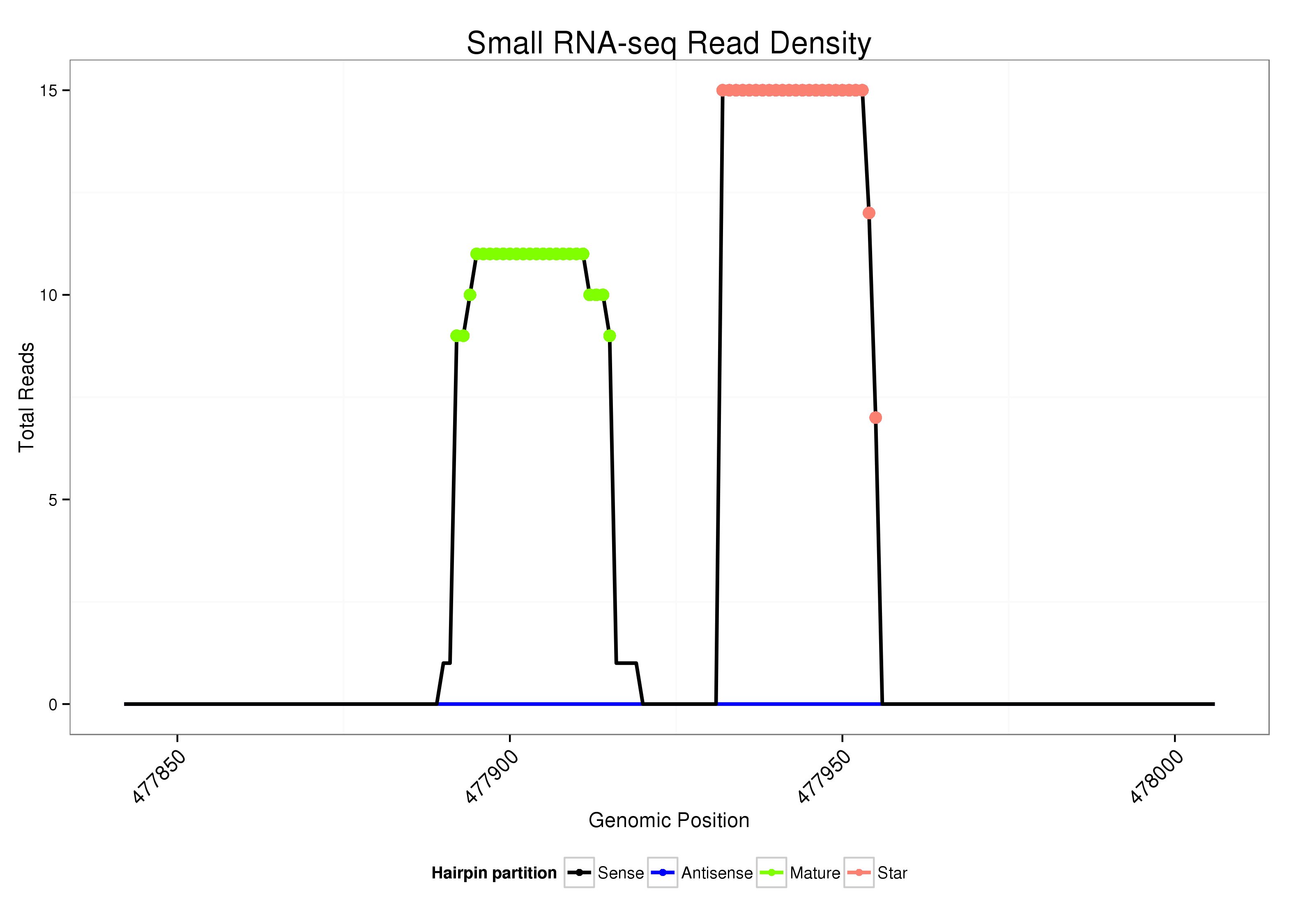
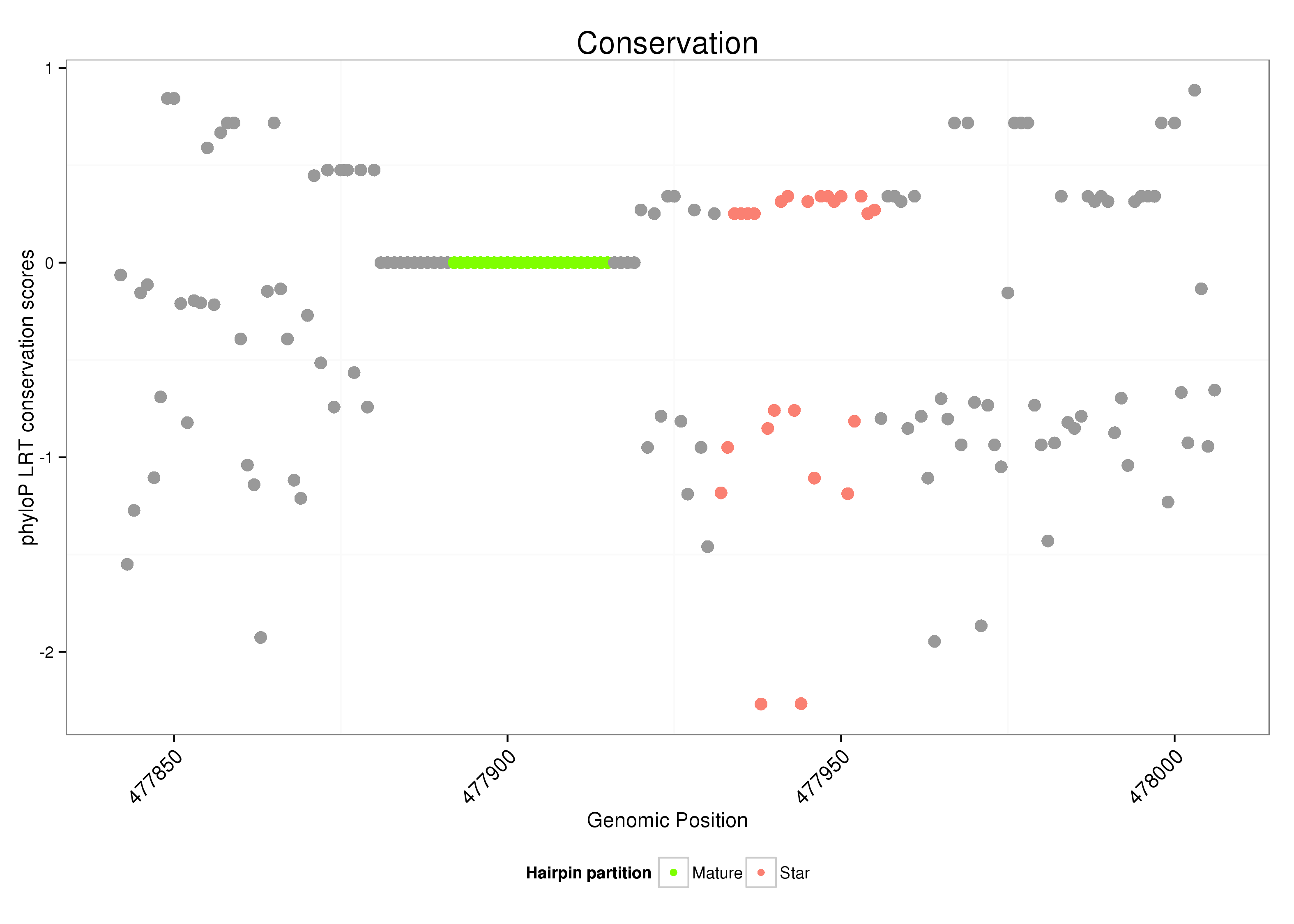
ID:dsi_90 |
Coordinate:3L:477892-477956 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
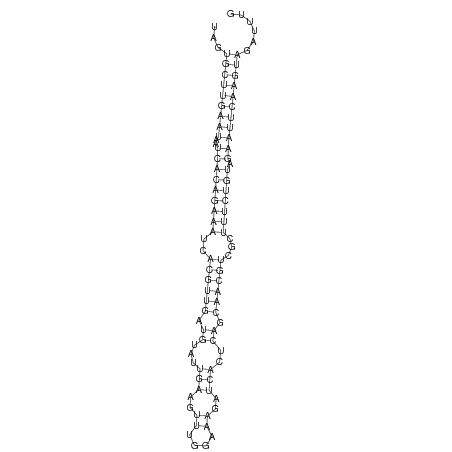 |
 |
 |
 |
| -23.4 | -23.1 | -23.0 |
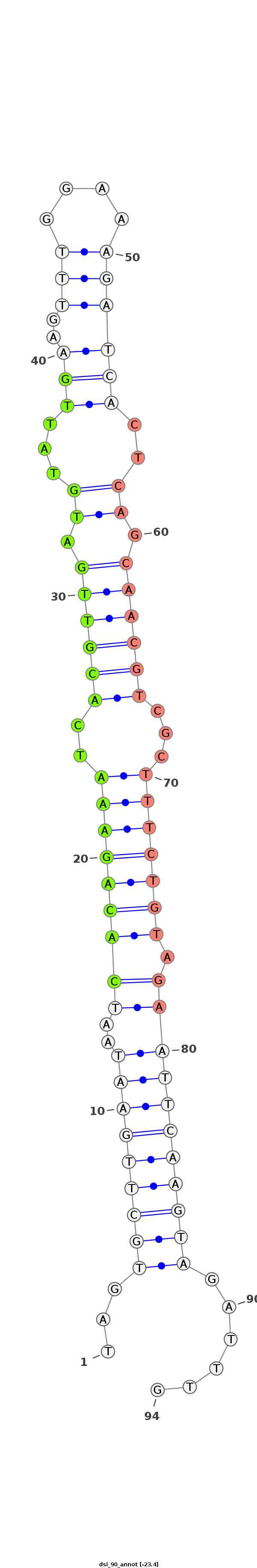 |
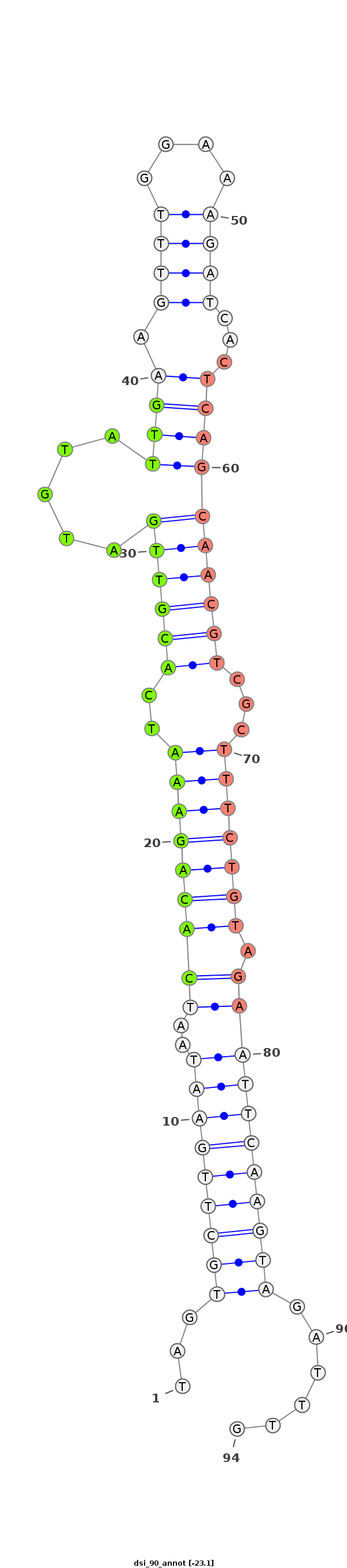 |
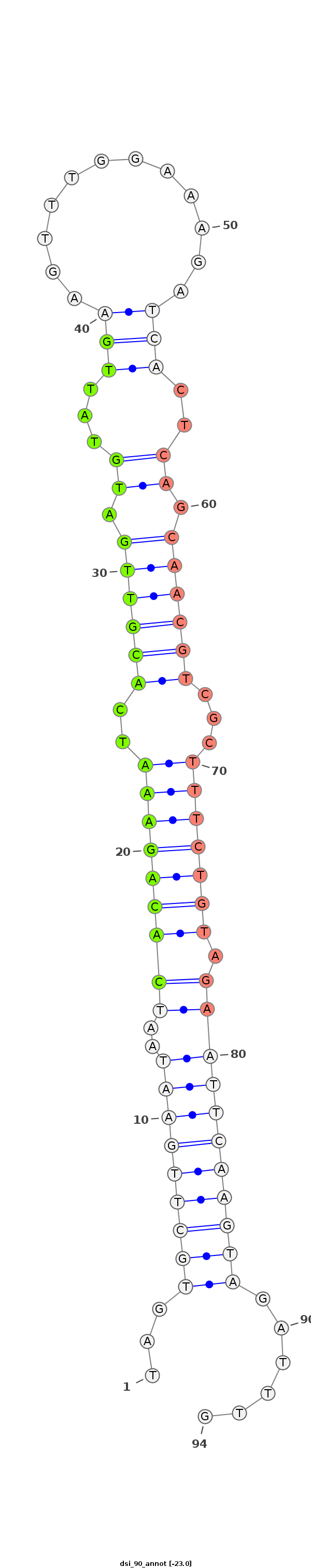 |
intergenic
No Repeatable elements found
|
AAGGGGCTAAATGTTACATTCATTTTGACATTTAGTAGTGCTTGAATAATCACAGAAATCACGTTGATGTATTGAAGTTTGGAAAGATCACTCAGCAACGTCGCTTTCTGTAGAATTCAAGTAGATTTGACTTCATTACCTTTTCACAGATTCTAAAGAATTATA
***********************************...(((((((((..(((((((((..((((((.((...(((..((....))..)))..)).))))))...))))))).)))))))))))......************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR618934 Ovary |
M024 male body |
M025 embryo |
O002 Head |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................CACAGAAATCACGTTGATGTATTG........................................................................................... | 24 | 0 | 1 | 8.00 | 8 | 5 | 0 | 1 | 1 | 1 |
| ..........................................................................................CTCAGCAACGTCGCTTTCTGTAGA................................................... | 24 | 0 | 1 | 7.00 | 7 | 5 | 2 | 0 | 0 | 0 |
| ..........................................................................................CTCAGCAACGTCGCTTTCTGTAG.................................................... | 23 | 0 | 1 | 5.00 | 5 | 3 | 2 | 0 | 0 | 0 |
| ..........................................................................................CTCAGCAACGTCGCTTTCTGTA..................................................... | 22 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ....................................................CAGAAATCACGTTGATGTATT............................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................AGAAATCACGTTGATGTATTGAAGT....................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................ATCACAGAAATCACGTTGATGT............................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................CTCAGCGACGTCGCCTTGTGT...................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................AGAAGGGCTTGAATAATCGC................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 |
|
TATAATTCTTTAGAATCTGTGAAAAGGTAATGAAGTCAAATCTACTTGAATTCTACAGAAAGCGACGTTGCTGAGTGATCTTTCCAAACTTCAATACATCAACGTGATTTCTGTGATTATTCAAGCACTACTAAATGTCAAAATGAATGTAACATTTAGCCCCTT
************************************......(((((((((((.(((((((...((((((.((..(((..((....))..)))...)).))))))..)))))))))..)))))))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|---|---|---|---|---|
| ...............TCTGTCAAAAGGTAACG..................................................................................................................................... | 17 | 2 | 9 | 0.44 | 4 |
| ..............ATCTGTCAAAAGGTAACGCA................................................................................................................................... | 20 | 3 | 8 | 0.25 | 2 |
| ..............ATCTGTCAAAAGGTA........................................................................................................................................ | 15 | 1 | 15 | 0.20 | 3 |
| .................TGTGAAAAGTTAATGAAGTAG............................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 |
| ...............TCTGTCAAAAGGTAACGCAG.................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 |
| ...............TCTGTTAAAAGGTAACG..................................................................................................................................... | 17 | 2 | 12 | 0.08 | 1 |
| .............................................................GCGACGTCGCTAAGCGATC..................................................................................... | 19 | 3 | 13 | 0.08 | 1 |
| .........................................................GAAAGCGACGTCGCT............................................................................................. | 15 | 1 | 20 | 0.05 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | 3L:477842-478006 + | dsi_90 | AAGGGGC-----------------TAAATGTTACATTCATTTT----------------------------------------------------------GACATTTAGTAGTGCTTGAATAATCACAGAAATCACGTTGATGTATTGAAGTTTGGAAAGATCACTCAGCAACGTCGCTTTCTGTAGAATTCA-----------------------------AGTAGATTTGA----------CTT---------------CAT-TAC---------CTTTTCACAGATTCTAAAGAATTATA |
| droSec2 | scaffold_2:852355-852496 + | TAGGTGC-----------------TAAATGTTATATTCATTTT----------------------------------------------------------GACGTTTATTAGTGCTT---------------------TGATCTATTGAAGTTTGGAAAGATCCCTCGGCAACGTCTCTTTCTGAAGAATTCA-----------------------------AGTAAATCTGA----------TTT---------------GAT-TAC---------CTTGTCACAGTCTCTAAAGAATTC-- | |
| dm3 | chr3L:824561-824716 + | TATGTTG-----------------CTCATTTTACATTCATTTT----------------------------------------------------------GACATTGAACAGTGCTTAAATAATCATAGAAATCAC-------TATTGAAGTTCGGAAAGATCCCTCAGCAAC-----TGTGTGAAAAATTC-------------------------GTTTAAGTAGATTTCA----------TTT---------------GGT-TTC---------ATTTTAGGAGTTTCCAAAGAATT-TC | |
| droEre2 | scaffold_4784:840100-840255 + | T------------------------------TATATACCTTTT----------------------------------------------------------AAC-------------------------------------------------TCGAAAATACGCACCGGCCGCGTCGCGTTCTGAAGAATTCGCGTGCCATCTCTGCTTGACTATTCATTTGAAGCGCTCTCTCTAAATTTGTCACTTCTATGAAATCTCTGAT-TGC-----------TTCTACAGATTCTAAAGAACTCGC | |
| droYak3 | 3L:820097-820223 + | TGTGTTC-----------------TAATTG---TACATTTTTC----------------------------------------------------------AGT-----------------TATTCTAAG--------------------------GAAT-----------GATGTTCCTTTCTCTA--GTTCA-----------------------------AGTTGCTTTGG----------CACTACTATCAAATCTCTGAT-TGTTTCTATGCTGCTTTCACAGTTGCTAAATAATT--- | |
| droEug1 | scf7180000409184:858498-858574 + | ACTTTGA-----------------TA---------------------------TTATTTCGGAAATAAGAAACACTAATTATTGTATAATCCACTCATTACTTCGCTGATAATT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTCTA | |
| droKik1 | scf7180000302383:193742-193859 + | TGGGGATGATCTTGAGGCTTTAGATATACATAATATCAAATCTTATATATGTATTTGTTTTGAAAT-GGAAAT------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGA----------TGATTTTATAGAATTTCTAATCTATATTTTTG-------------------ATACTTAAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 12/06/2013 at 02:30 PM