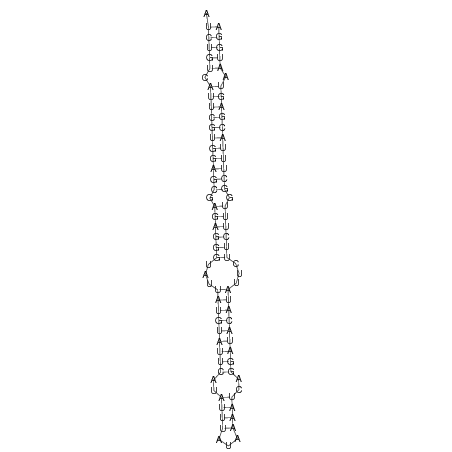
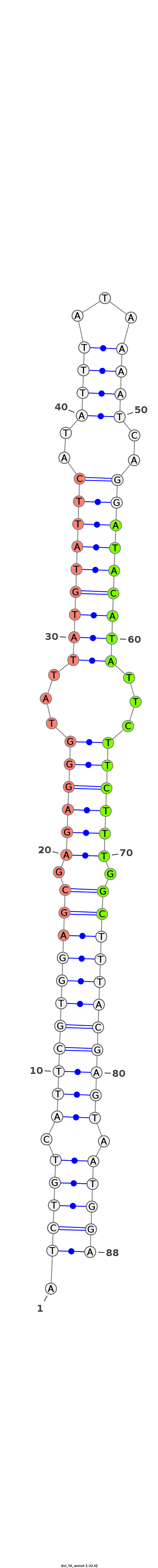
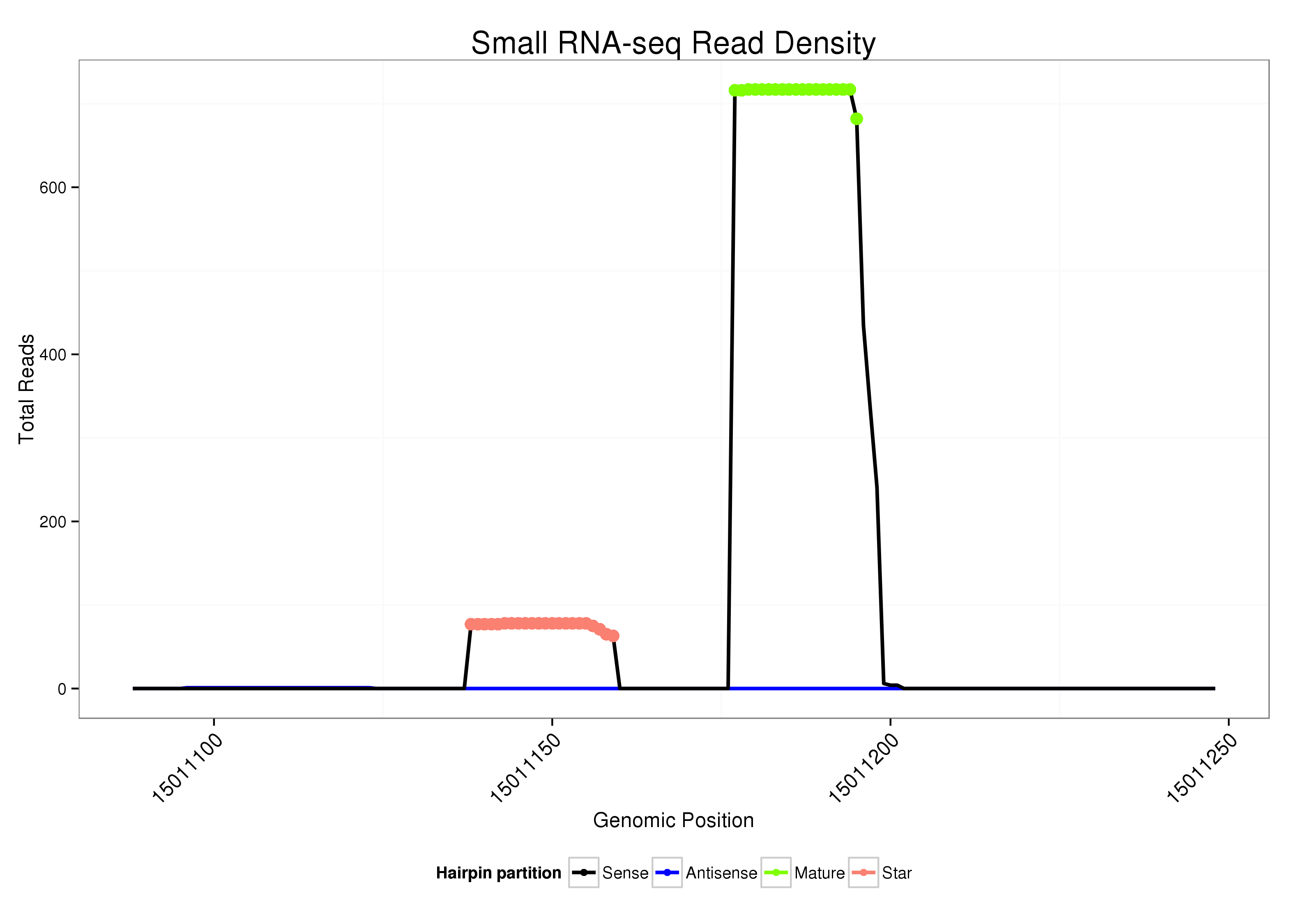
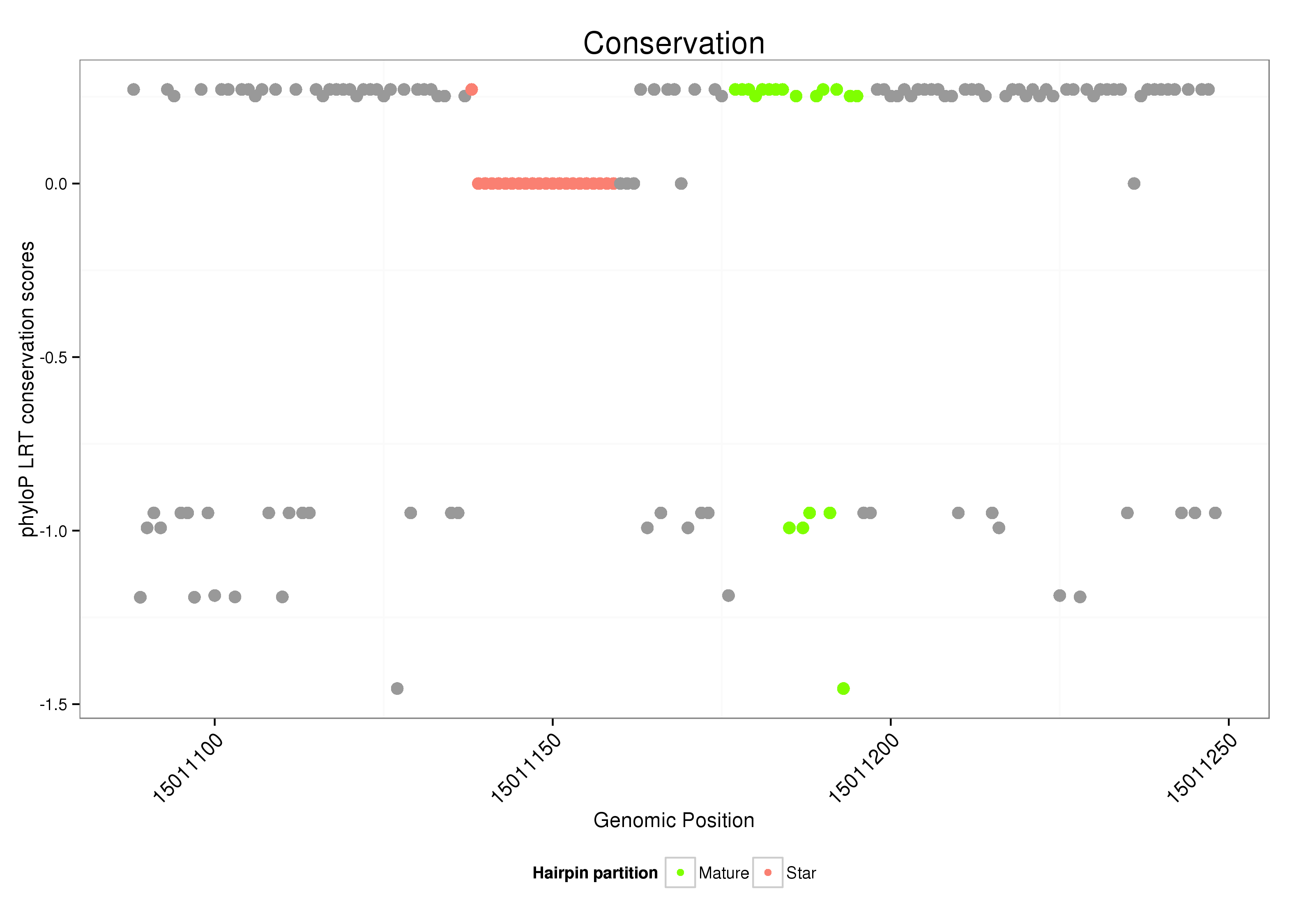
ID:dsi_56 |
Coordinate:X:15011138-15011198 + |
Confidence:Known Ortholog |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -32.1 | -31.9 | -31.8 |
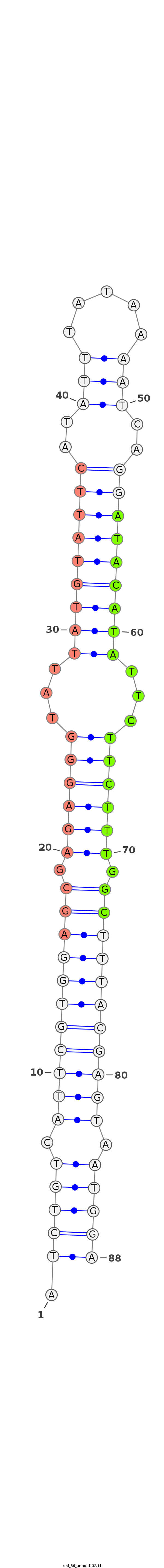 |
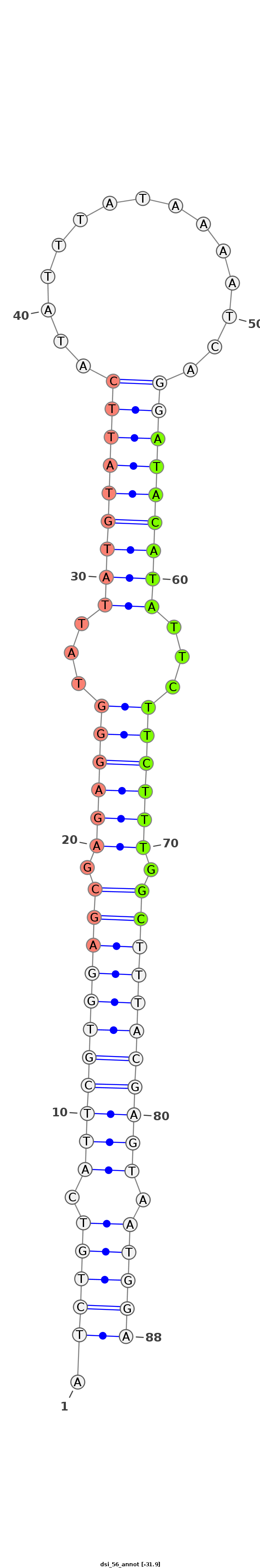 |
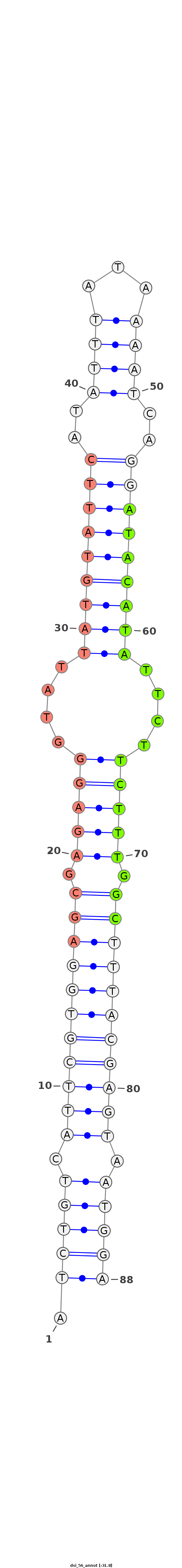 |
intergenic
No Repeatable elements found
|
AAAGTACGCCATGAATATCTTTAAATTTCATTTGTATCTGTCATTCGTGGAGCGAGAGGGTATTATGTATTCATATTTATAAAATCAGGATACATATTCTTCTTTGGCTTTACGAGTAATGGATATCGTCAACAGTGGAATACTAATGTGAAAAATATAAA
***********************************.(((((.(((((((((((.((((((...(((((((((..((((...))))..)))))))))...)))))).))))))))))).)))))************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
M024 male body |
SRR553488 Ovary |
O002 Head |
SRR618934 Ovary |
SRR553486 Ovary |
M023 head |
SRR553485 Ovary |
SRR553487 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................ATACATATTCTTCTTTGGC..................................................... | 19 | 0 | 1 | 247.00 | 247 | 243 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ATACATATTCTTCTTTGGCTTT.................................................. | 22 | 0 | 1 | 234.00 | 234 | 184 | 18 | 11 | 5 | 2 | 5 | 0 | 3 | 6 |
| .........................................................................................ATACATATTCTTCTTTGGCT.................................................... | 20 | 0 | 1 | 99.00 | 99 | 73 | 22 | 0 | 0 | 0 | 0 | 3 | 1 | 0 |
| .........................................................................................ATACATATTCTTCTTTGGCTT................................................... | 21 | 0 | 1 | 95.00 | 95 | 34 | 50 | 2 | 1 | 2 | 4 | 0 | 2 | 0 |
| ..................................................AGCGAGAGGGTATTATGTATTC......................................................................................... | 22 | 0 | 1 | 62.00 | 62 | 34 | 21 | 0 | 1 | 4 | 0 | 2 | 0 | 0 |
| .........................................................................................ATACATATTCTTCTTTGG...................................................... | 18 | 0 | 1 | 35.00 | 35 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGCGAGAGGGTATTATGTAT........................................................................................... | 20 | 0 | 1 | 6.00 | 6 | 4 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................AGCGAGAGGGTATTATGTA............................................................................................ | 19 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ATACATATTCTTCTTTGGCTTTACG............................................... | 25 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ATACATATTCTTCTTTGGCC.................................................... | 20 | 1 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGCGAGAGGGTATTATGT............................................................................................. | 18 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................ATACATATTCTTCTTTGGCTTTA................................................. | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGCGAGAGGGTATTATGTATT.......................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................ATACATATTCTTCTTTGGCTC................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................GAGGGTATTATGTATTC......................................................................................... | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................ACATATTCTTCTTTGGCTTT.................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGCGAGAGGGTATTATGTATTCAA....................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGCGAGACGGTATTATGTATTC......................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ATACATATTCTTCTTTGGCTTTAAA............................................... | 25 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................ATACATATTCTTCTTTGGCTTTT................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ATACATATTCTTCTTTGGCTTC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ATAAATATTCTTCTTTGGCT.................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTTATATTTTTCACATTAGTATTCCACTGTTGACGATATCCATTACTCGTAAAGCCAAAGAAGAATATGTATCCTGATTTTATAAATATGAATACATAATACCCTCTCGCTCCACGAATGACAGATACAAATGAAATTTAAAGATATTCATGGCGTACTTT
**************************************(((((.(((((((((((.((((((...(((((((((..((((...))))..)))))))))...)))))).))))))))))).))))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 Ovary |
|---|---|---|---|---|---|---|
| .....................................................................................................................ACGACAGATACAAATGAAATTTAAAGA................. | 27 | 1 | 1 | 1.00 | 1 | 0 |
| ...................................................................TGTATCCTGATTTTATAAATGTGAA..................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 |
| ........TTTCACATTAGTATTCCACTGTTGACGA............................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................................................ACCCTGTCGCACCACGA............................................ | 17 | 2 | 6 | 0.50 | 3 | 1 |
| ...................AATTCTACTGTTGACGA............................................................................................................................. | 17 | 2 | 9 | 0.11 | 1 | 0 |
| ...........................................................GAAGAATGTGTATCCGG..................................................................................... | 17 | 2 | 10 | 0.10 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | X:15011088-15011248 + | dsi_56 | AAAGTACGCCATGAATATCTTTAAATTT-CATTT-GTATC--TGTCATTCGTGGAGCGAGAGGGTATTATGTATTCATATTTATAAAATCA-GGATACATATTCTTCTTTGGCTTTACGAGTAATGGAT--ATCGTCAACAGTGGAATACTAATGTGAAA--AAT-ATA-AA |
| droSec2 | scaffold_8:1727757-1727918 + | dse_85 | AAAGTACGCCTCGAATATCTTTTAATTT-CATTT-GTATC--TCTCATTCGTGGAGCGAGAGGGTATTATGTATTCATATTTATAAAATCA-GGATACATATTCTTCTTTGGCTTTACGAGTAATGGAT--ATCGTCAACAGTGGAATACTAATGTGAAA--AAT-ATAAAA |
| dm3 | chrX:19445325-19445465 + | dme-mir-9369 | AAAGTG---------------------------T-GTATCTCTCTCATTCGTTTAGCGAGAGGGTATTATGTATTCATATTTGTAATATCA-TGATACATATTCTCCTTTCGCTCTACGAGTAATGGATATATCGTCAACAGTGAAATA-TAATAATAAAATAATGATA-AA |
| droEre2 | scaffold_4690:9682065-9682202 + | ACTAAACATATTTAAGATCTCTGGACCTACATTTAGTATC--TGTTATTCGCAGA------------------------TATGTA-TACTATGTATACATATACACCTCTCGCCCTACGAGTAATGGGT--ATCAACAACAGTGTAAGACTAATA-GAAA--AAC-ACA-AG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 12/06/2013 at 02:29 PM