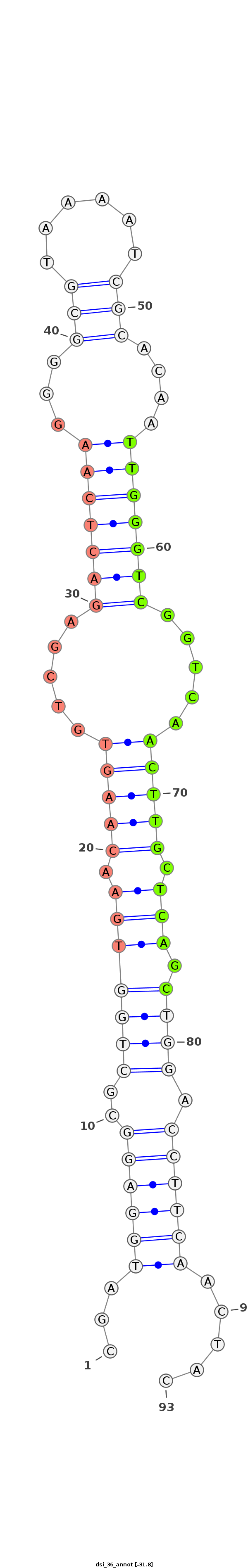
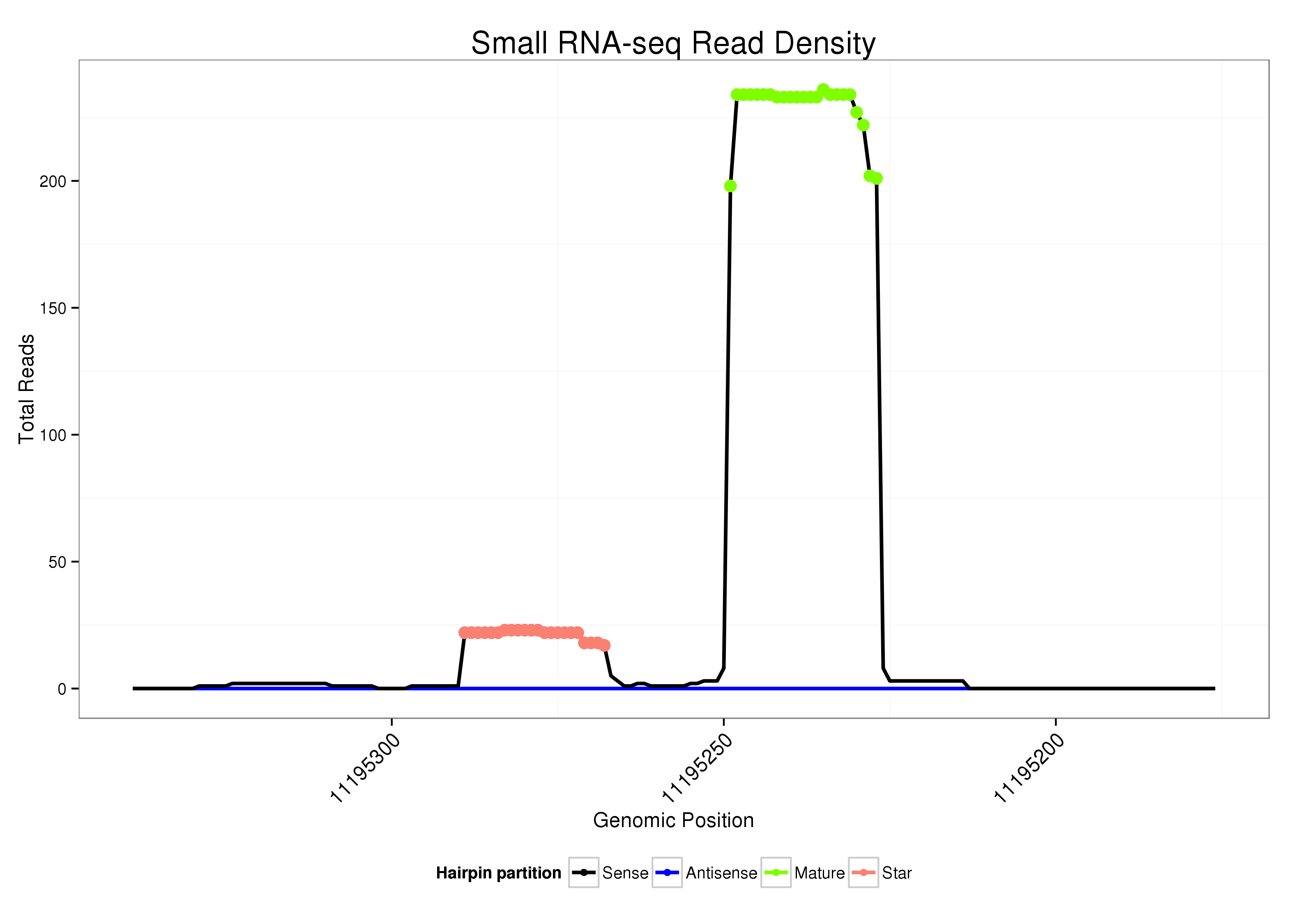
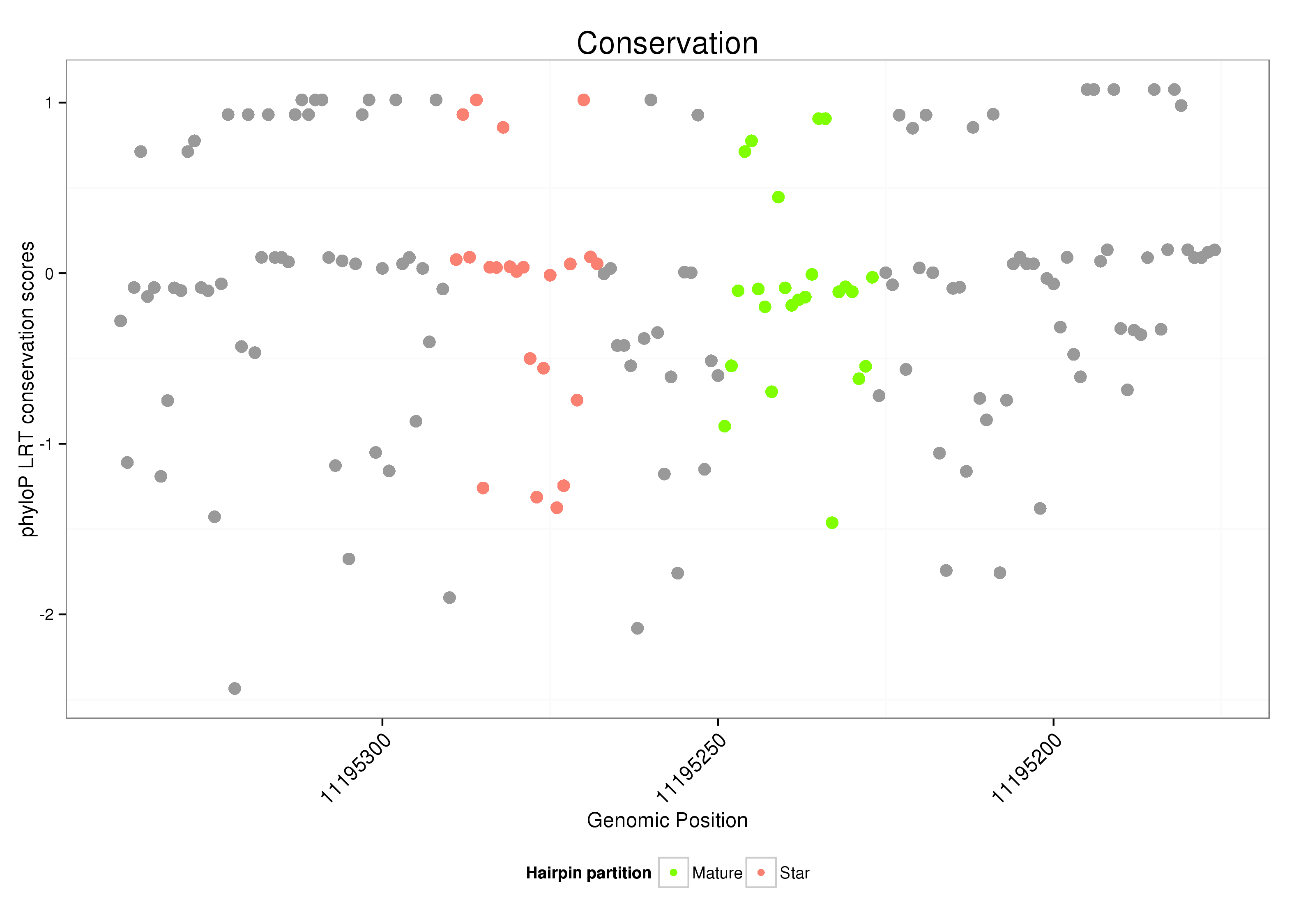
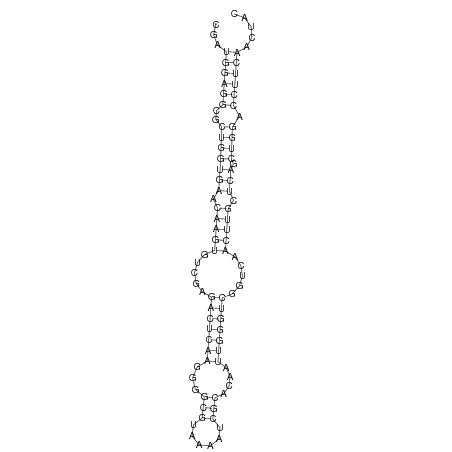| ..........................................................................................TTGGGTCGGTCAACTTGCTCAGC................................................... |
23 |
0 |
1 |
155.00 |
155 |
90 |
62 |
2 |
0 |
1 |
0 |
0 |
| ...........................................................................................TGGGTCGGTCAACTTGCTCAGC................................................... |
22 |
0 |
1 |
35.00 |
35 |
25 |
10 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCTCAGT................................................... |
23 |
1 |
1 |
23.00 |
23 |
8 |
15 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCTCA..................................................... |
21 |
0 |
1 |
19.00 |
19 |
11 |
8 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TGAACAAGTGTCGAGACTCAAG............................................................................................ |
22 |
0 |
1 |
12.00 |
12 |
5 |
7 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCT....................................................... |
19 |
0 |
1 |
7.00 |
7 |
3 |
4 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCTCAGG................................................... |
23 |
1 |
1 |
6.00 |
6 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCTCAGA................................................... |
23 |
1 |
1 |
6.00 |
6 |
2 |
4 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCTCAGCT.................................................. |
24 |
0 |
1 |
5.00 |
5 |
4 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TGAACAAGTGTCGAGACT................................................................................................ |
18 |
0 |
1 |
4.00 |
4 |
1 |
3 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCTC...................................................... |
20 |
0 |
1 |
4.00 |
4 |
1 |
2 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................................TTGCTCAGCTGGACCTTCAACT...................................... |
22 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................ATTGGGTCGGTCAACTTGCTCAGC................................................... |
24 |
0 |
1 |
3.00 |
3 |
0 |
2 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................ATTGGGTCGGTCAACTTGCTCAGT................................................... |
24 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TGAACAAGTGTCGAGACTCAAGGG.......................................................................................... |
24 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TGAACAAGTGTCGAGACTCAAGG........................................................................................... |
23 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................TGGGTCGGTCAACTTGCTCAGT................................................... |
22 |
1 |
1 |
2.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................GTAACGATGGCGGCGCTGT.................................................................................................................. |
19 |
2 |
2 |
1.50 |
3 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..........CTACCGCAGGCACTAAGAGA...................................................................................................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ......................................................................................ACAATTGGGTCGGTCAACT........................................................... |
19 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................GTAAAATCGCACAATTGGGTC................................................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................ATTGGGTCGGTCAACTTGCTCAG.................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................GCACAATTGGGTCGGTCAACT........................................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ........................................................AGTGTCGAGACTCAAGGGGCGT...................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ...........................................................................................TGGGTCGGTCAACTTGCTCA..................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TAAACAAGTGTCGAGACTCAAGGGGC........................................................................................ |
26 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TGAACAAGTGTCGAGACTCAAGGT.......................................................................................... |
24 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACGTGCTCAGC................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................GTAACGATGGCGGCGCTGG.................................................................................................................. |
19 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................................................TGGGTCGGTCAACTTGCTT...................................................... |
19 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..........................................GGCGCTGGTGAACAAGTGTC...................................................................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCTCAA.................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............GCAGGCACTAAGAGAAGTAACG............................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .........................................................................................ATTGGGTCGGTCAACTTGCTC...................................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................TTGGGTCGGTCAACTTGCTCAGCC.................................................. |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TGAACAAGTGTCGAGACTCAA............................................................................................. |
21 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..............................................CTGGTGAACAAGAATCGAGC.................................................................................................. |
20 |
3 |
5 |
0.80 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................CGTAACGATGGCGGCGCTG................................................................................................................... |
19 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................TAACGATGGCGGCGCTGT.................................................................................................................. |
18 |
2 |
5 |
0.20 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................CGTTGGAGGCGCTGGT................................................................................................................. |
16 |
1 |
8 |
0.13 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ...............................GTAACGATGGCGGCG...................................................................................................................... |
15 |
1 |
12 |
0.08 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................TAACGATCGAGACTCTGG.................................................................................................................. |
18 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |