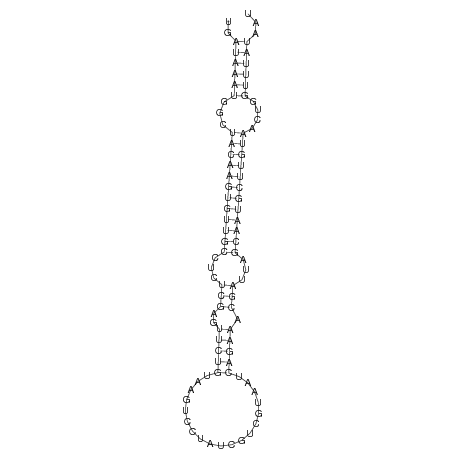
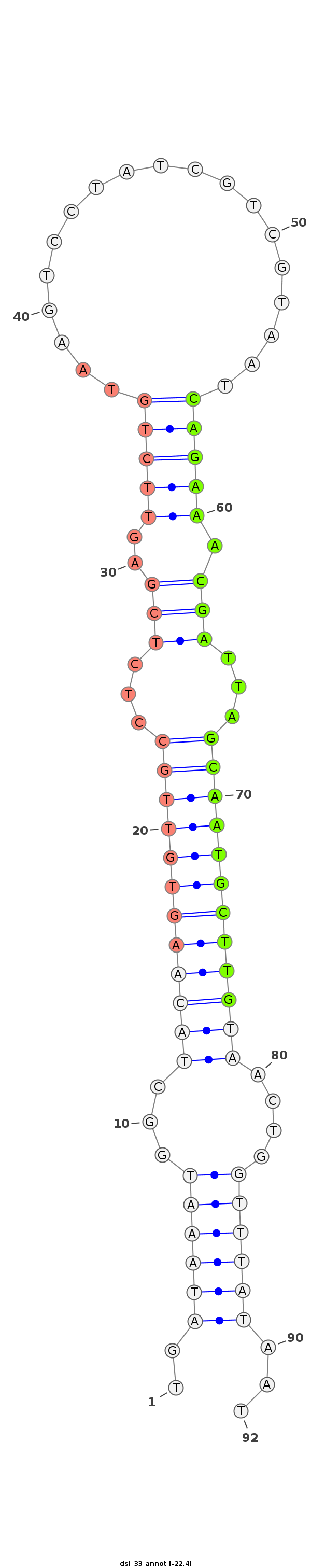
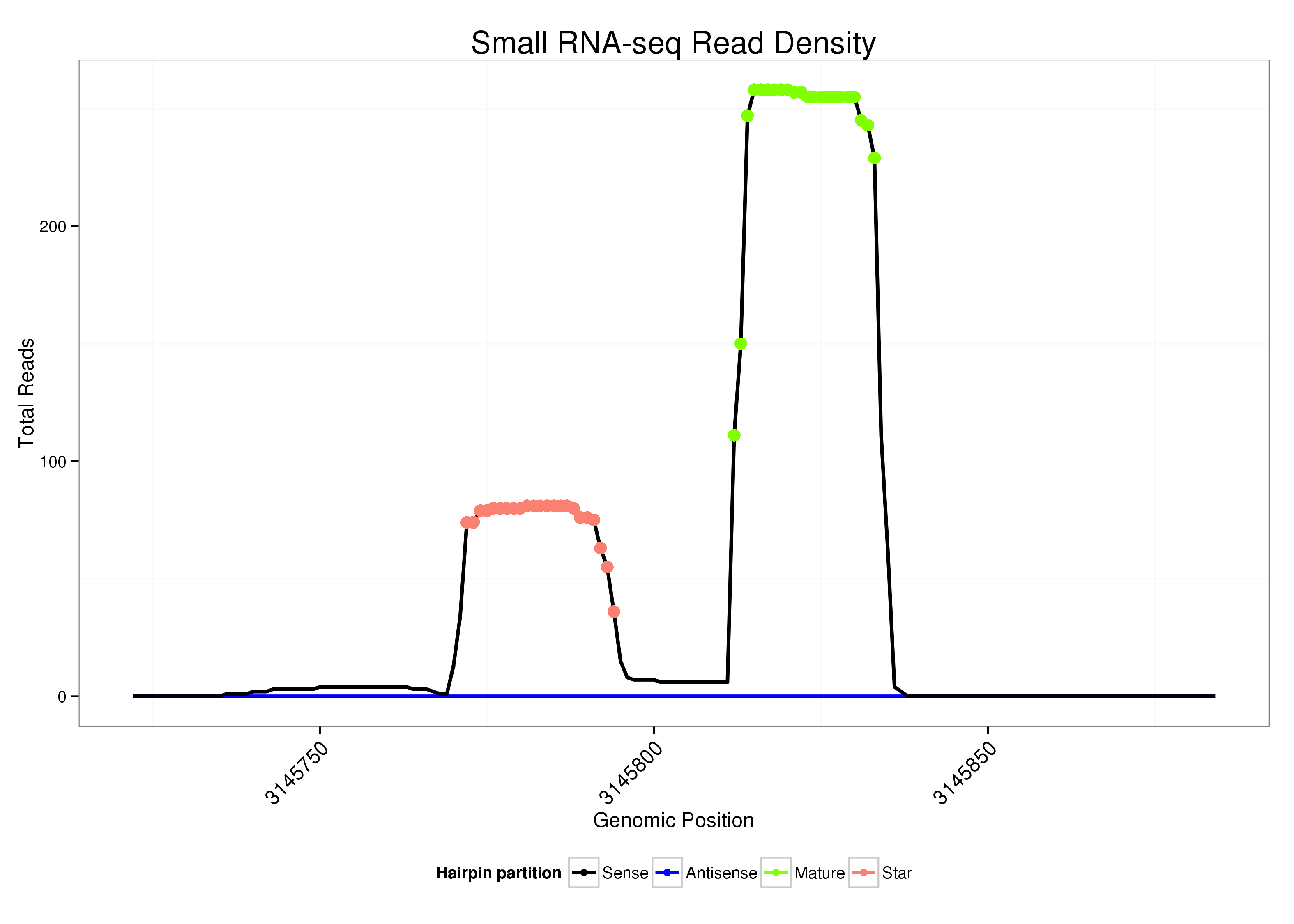
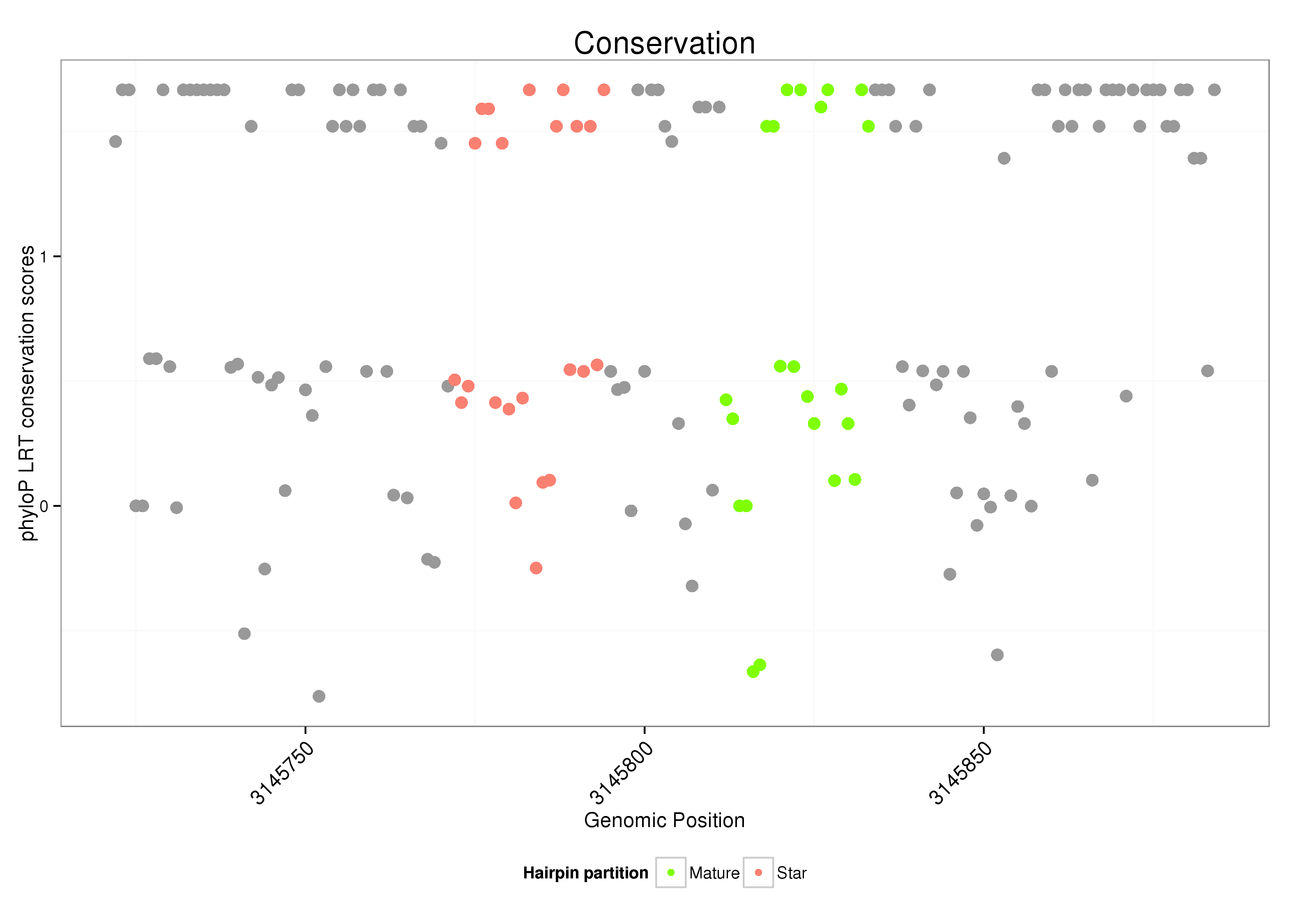
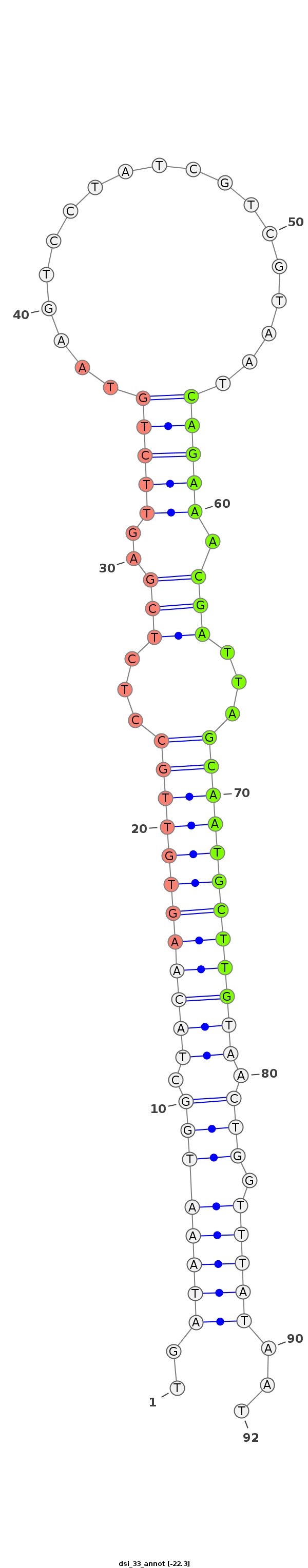
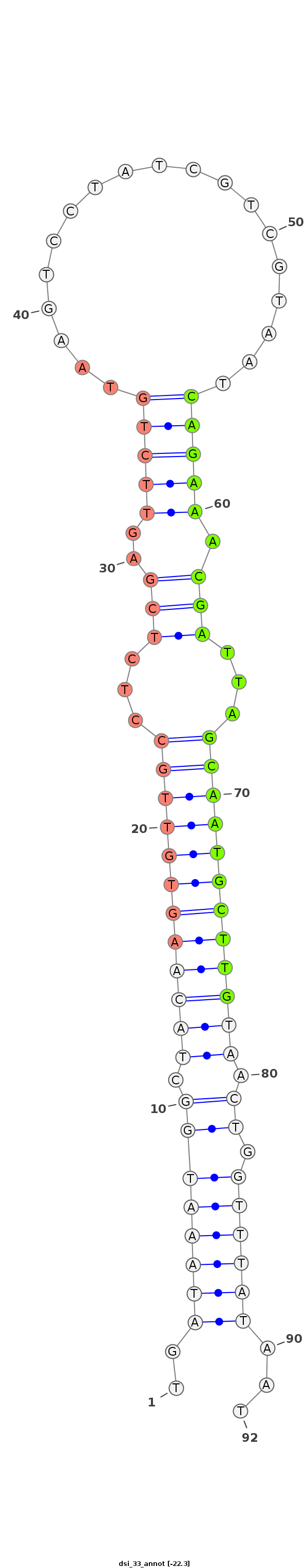
| ..........................................................................................CAGAAACGATTAGCAATGCTTG................................................... |
22 |
0 |
2 |
85.50 |
171 |
110 |
4 |
33 |
4 |
2 |
0 |
2 |
0 |
1 |
15 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTTGTA................................................. |
22 |
0 |
2 |
54.00 |
108 |
3 |
3 |
1 |
1 |
2 |
42 |
26 |
0 |
13 |
0 |
14 |
3 |
| ............................................................................................GAAACGATTAGCAATGCTTGT.................................................. |
21 |
0 |
2 |
33.50 |
67 |
2 |
38 |
1 |
21 |
4 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ...........................................................................................AGAAACGATTAGCAATGCTTG................................................... |
21 |
0 |
2 |
22.00 |
44 |
23 |
7 |
5 |
1 |
2 |
0 |
1 |
0 |
0 |
5 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTGTA.......................................................................................... |
23 |
0 |
2 |
21.00 |
42 |
23 |
0 |
6 |
2 |
0 |
0 |
1 |
0 |
5 |
3 |
1 |
1 |
| ...........................................................................................AGAAACGATTAGCAATGCTTGT.................................................. |
22 |
0 |
2 |
13.50 |
27 |
2 |
9 |
0 |
5 |
2 |
0 |
0 |
6 |
0 |
0 |
1 |
2 |
| ..........................................................................................CAGAAACGATTAGCAATGCTT.................................................... |
21 |
0 |
2 |
11.00 |
22 |
5 |
1 |
0 |
1 |
10 |
0 |
0 |
4 |
1 |
0 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTGT........................................................................................... |
22 |
0 |
2 |
10.00 |
20 |
5 |
6 |
0 |
4 |
2 |
0 |
0 |
0 |
0 |
2 |
0 |
1 |
| .................................................AAGTGTTGCCTCTCGAGTTCTGT........................................................................................... |
23 |
0 |
2 |
8.50 |
17 |
2 |
4 |
2 |
6 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................CAGAAACGATTAGCAATGC...................................................... |
19 |
0 |
2 |
7.00 |
14 |
0 |
0 |
0 |
0 |
4 |
0 |
0 |
10 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTTG................................................... |
20 |
0 |
2 |
7.00 |
14 |
8 |
0 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
| .................................................AAGTGTTGCCTCTCGAGTTCT............................................................................................. |
21 |
0 |
2 |
6.00 |
12 |
0 |
2 |
0 |
2 |
0 |
2 |
1 |
2 |
2 |
0 |
1 |
0 |
| ....................................................TGTTGCCTCTCGAGTTCTGTAAG........................................................................................ |
23 |
0 |
2 |
5.50 |
11 |
6 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTTGTAAA............................................... |
24 |
1 |
2 |
5.00 |
10 |
0 |
3 |
0 |
0 |
0 |
5 |
1 |
0 |
0 |
0 |
1 |
0 |
| ................................................CAAGTGTTGCCTCTCGAGTTCT............................................................................................. |
22 |
0 |
2 |
5.00 |
10 |
1 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
2 |
1 |
2 |
0 |
| ................................................CAAGTGTTGCCTCTCGAGTTCTG............................................................................................ |
23 |
0 |
2 |
4.50 |
9 |
2 |
0 |
3 |
2 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
| .............................................................................................AAACGATTAGCAATGCTTGTAAA............................................... |
23 |
1 |
2 |
4.50 |
9 |
0 |
2 |
0 |
0 |
0 |
4 |
3 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTG............................................................................................ |
21 |
0 |
2 |
4.50 |
9 |
4 |
0 |
0 |
0 |
2 |
0 |
0 |
2 |
0 |
1 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTGTAA......................................................................................... |
24 |
0 |
2 |
4.50 |
9 |
0 |
0 |
7 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .............................................................................................AAACGATTAGCAATGCTTGTA................................................. |
21 |
0 |
2 |
4.00 |
8 |
0 |
1 |
1 |
0 |
0 |
2 |
3 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................................CAGAAACGATTAGCAATGCTGG................................................... |
22 |
1 |
2 |
4.00 |
8 |
6 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .............................................................................................AAACGATTAGCAATGCTTG................................................... |
19 |
0 |
2 |
3.50 |
7 |
5 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGTGTTGCCTCTCGAGTTCTGTAA......................................................................................... |
25 |
0 |
2 |
3.50 |
7 |
0 |
0 |
2 |
0 |
0 |
1 |
0 |
0 |
2 |
0 |
2 |
0 |
| ...........................................................................................AGAAACGATTAGCAATGC...................................................... |
18 |
0 |
2 |
3.00 |
6 |
0 |
0 |
0 |
0 |
4 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| .............................................................................................AAACGATTAGCAATGCTTGT.................................................. |
20 |
0 |
2 |
3.00 |
6 |
0 |
5 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGTGTTGCCTCTCGAGT................................................................................................ |
18 |
0 |
2 |
3.00 |
6 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
| ..........................................................................................CAGAAACGATTAGCAATGCT..................................................... |
20 |
0 |
2 |
2.50 |
5 |
0 |
0 |
1 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGTGTTGCCTCTCGAGTTCTG............................................................................................ |
22 |
0 |
2 |
2.50 |
5 |
2 |
0 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................TCCTATCGTCGTAATCAGAAACGATT.............................................................. |
26 |
0 |
1 |
2.00 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTTGTAAC............................................... |
24 |
0 |
2 |
2.00 |
4 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| .......................................................................TAAGTCCTATCGTCGTAATCA....................................................................... |
21 |
0 |
1 |
2.00 |
2 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTGTAAA........................................................................................ |
25 |
1 |
2 |
2.00 |
4 |
0 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTT.................................................... |
19 |
0 |
2 |
2.00 |
4 |
0 |
0 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................AGAAACGATTAGCAATGCTT.................................................... |
20 |
0 |
2 |
1.50 |
3 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTTGTAA................................................ |
23 |
0 |
2 |
1.50 |
3 |
0 |
1 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ................................................CAAGTGTTGCCTCTCGAGTTC.............................................................................................. |
21 |
0 |
2 |
1.50 |
3 |
0 |
0 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTGGA.......................................................................................... |
23 |
1 |
2 |
1.00 |
2 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................CAAGTGTTGCCTCTCGAGT................................................................................................ |
19 |
0 |
2 |
1.00 |
2 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................TTGCCTCTCGAGTTCTGT........................................................................................... |
18 |
0 |
2 |
1.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................TCCTATCGTCGTAATCAGAAACGA................................................................ |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................GATTAGCAATGCTTG................................................... |
15 |
0 |
2 |
1.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................TGTTTAAGAATGTCTGATAAATGGC..................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ................................................CAAGTGTTTCCTCTCGAGTTCT............................................................................................. |
22 |
1 |
2 |
1.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................AAACGATTAGCAATGCTTGTAA................................................ |
22 |
0 |
2 |
1.00 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
| ............................................................................................GAGACGATTAGCAATGCTTGT.................................................. |
21 |
1 |
2 |
1.00 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
| ..............TATATGCTGTTTAAGAATGTCTGATAAA......................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...........................................................................................AGAAACGATTAGCAATGCTTGA.................................................. |
22 |
1 |
2 |
1.00 |
2 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
| ............................GAATGTCTGATAAATGGCTAC.................................................................................................................. |
21 |
0 |
2 |
1.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................TCCTATCGTCGTAATCAGAAAC.................................................................. |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTGG........................................................................................... |
22 |
1 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAATCCTTGTA................................................. |
22 |
1 |
2 |
1.00 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
| ..................TGCTGTTTAAGAATGTCTGATAAATGG...................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCT............................................................................................. |
20 |
0 |
2 |
1.00 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| ...........................................................TCTCGAGTTCTGTAAGTCCT.................................................................................... |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................CAAGTGTTGCCTCTCGAG................................................................................................. |
18 |
0 |
2 |
1.00 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| .............................................................................ATACCGTCGTTATCAGAA.................................................................... |
18 |
3 |
20 |
0.75 |
15 |
0 |
0 |
5 |
0 |
0 |
0 |
0 |
0 |
5 |
0 |
1 |
4 |
| ............................................................................................GAAACGATTAGCAATGCTTGTAGA............................................... |
24 |
2 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................CAGAAACGATAAGCAATGCTTG................................................... |
22 |
1 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................CATAAACGATTAGCAATGCTTG................................................... |
22 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGGGTTGCCTCTCGAGTTCTGT........................................................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGTGTTGCCTCTCGAGTTCTGA........................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTTGTG................................................. |
22 |
1 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGTTTTGCCTCTCGAGTTCTGTAA......................................................................................... |
24 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................CAAAAACGATTAGCAATGCTTG................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGATTTCTGTA.......................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTGTTGCCTCTCGAGTTCTGT........................................................................................... |
22 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAACCGATTAGCAACGCTTGT.................................................. |
21 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGTGTTGCCTCTCGAGTTCTGAA.......................................................................................... |
24 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................CTCTCGAGTTCTGTAA......................................................................................... |
16 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGTGTTGCCTCTCGAGTTCTGTAAA........................................................................................ |
26 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ....................................................TGTTGCCTCTCGAGTTCTGTAA......................................................................................... |
22 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................GTACGAATCCCAAAGTCATCGAAAGG............ |
26 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .................................................AAGTGCTGCCTCTCGCGGTCTGT........................................................................................... |
23 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTGTAAGT....................................................................................... |
26 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GGAACGATTAGCAATGCTTGT.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGTTTTGCCTCTCGAGTTCTGTA.......................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................AGAAATGATTAGCAATGCTTGT.................................................. |
22 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................CCCTCTCGAGTTCTGTATGT....................................................................................... |
20 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..............................................TACAAGTGTTGCCTCTCGAGTTCTGTA.......................................................................................... |
27 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| .................................................AAGTGTTGCCTCTCGAGTTCTGC........................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................AAACGATTAGCGATGCTTGT.................................................. |
20 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTTGTAAAA.............................................. |
25 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAACGATTAGCCATGCTTGT.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................GTTGCCTCTCGAGTTCTGT........................................................................................... |
19 |
0 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................TGCTGTGTAACAATGTCTGAAA........................................................................................................................... |
22 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAGCGATTAGCAATGCTTGT.................................................. |
21 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................CGGAAACGATTAGCAATGCTTG................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGTGTTTCCTCTCGAGTTCTG............................................................................................ |
22 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGCGTTCTGTA.......................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................CAAGTTTTGCCTCTCGAGTTCTG............................................................................................ |
23 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAACAATTAGCAATGCTTGT.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAATGATTAGCAATGCTTGT.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................TGCCTCTCGAGTTCTGTAAGTC...................................................................................... |
22 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..........................................................................................CAGAAGCGATTAGCAATGCTTG................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGTGTTGCCTCTCGAGTTCTGTAAG........................................................................................ |
25 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGTGTTGCCTCTCGCGTTCTGT........................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................TTGCCTCTCGAGTTCTGTAA......................................................................................... |
20 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAATGCTTGTAAAAA............................................. |
26 |
3 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GAAACGATTAGCAAT........................................................ |
15 |
0 |
3 |
0.33 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................CCTCTCGAGTTCTGT........................................................................................... |
15 |
0 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...........................................................TCTCGAGTTCTGTAAA........................................................................................ |
16 |
1 |
6 |
0.17 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..............................................................................TACCGTCGTTATCAGAACC.................................................................. |
19 |
3 |
6 |
0.17 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................TGCTGTTTAAGAACG.................................................................................................................................. |
15 |
1 |
6 |
0.17 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................ATACCGTCGTTATCAGAAA................................................................... |
19 |
3 |
13 |
0.15 |
2 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..............................................................................TACCGTCGTTATCAGAA.................................................................... |
17 |
2 |
7 |
0.14 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..............................................................................TACCGTCGTTATCAGAAAA.................................................................. |
19 |
3 |
17 |
0.12 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................TACCGTCGTTATCAGA..................................................................... |
16 |
2 |
19 |
0.11 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
| ..............................................................................TACCGTCGTTATCAGTAA................................................................... |
18 |
3 |
18 |
0.06 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................TACCGTCGTTATCAGAAC................................................................... |
18 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................AGTCGAATCCCTAAGT...................... |
16 |
2 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |