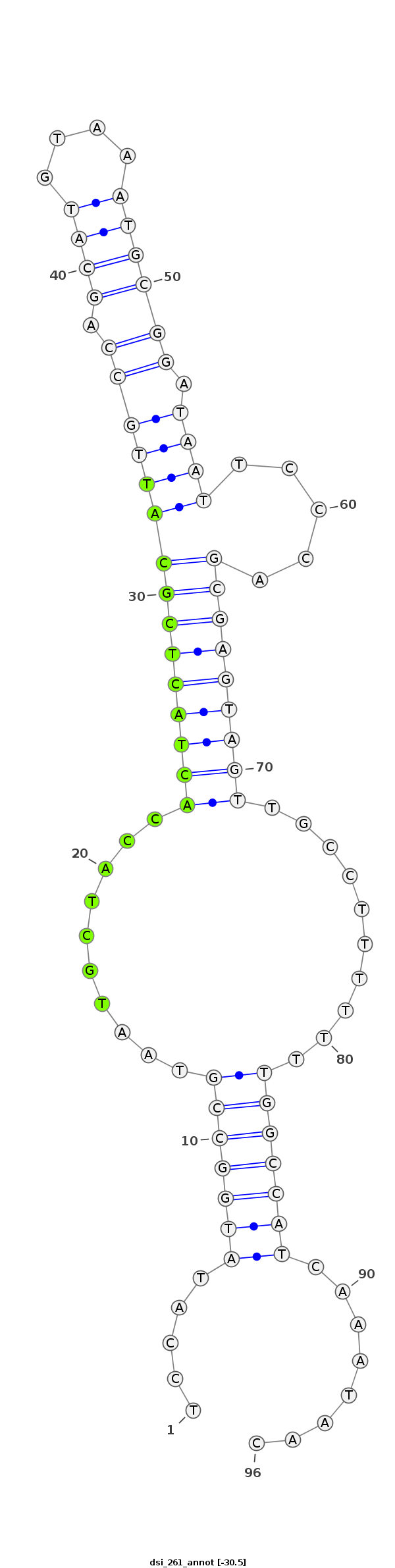
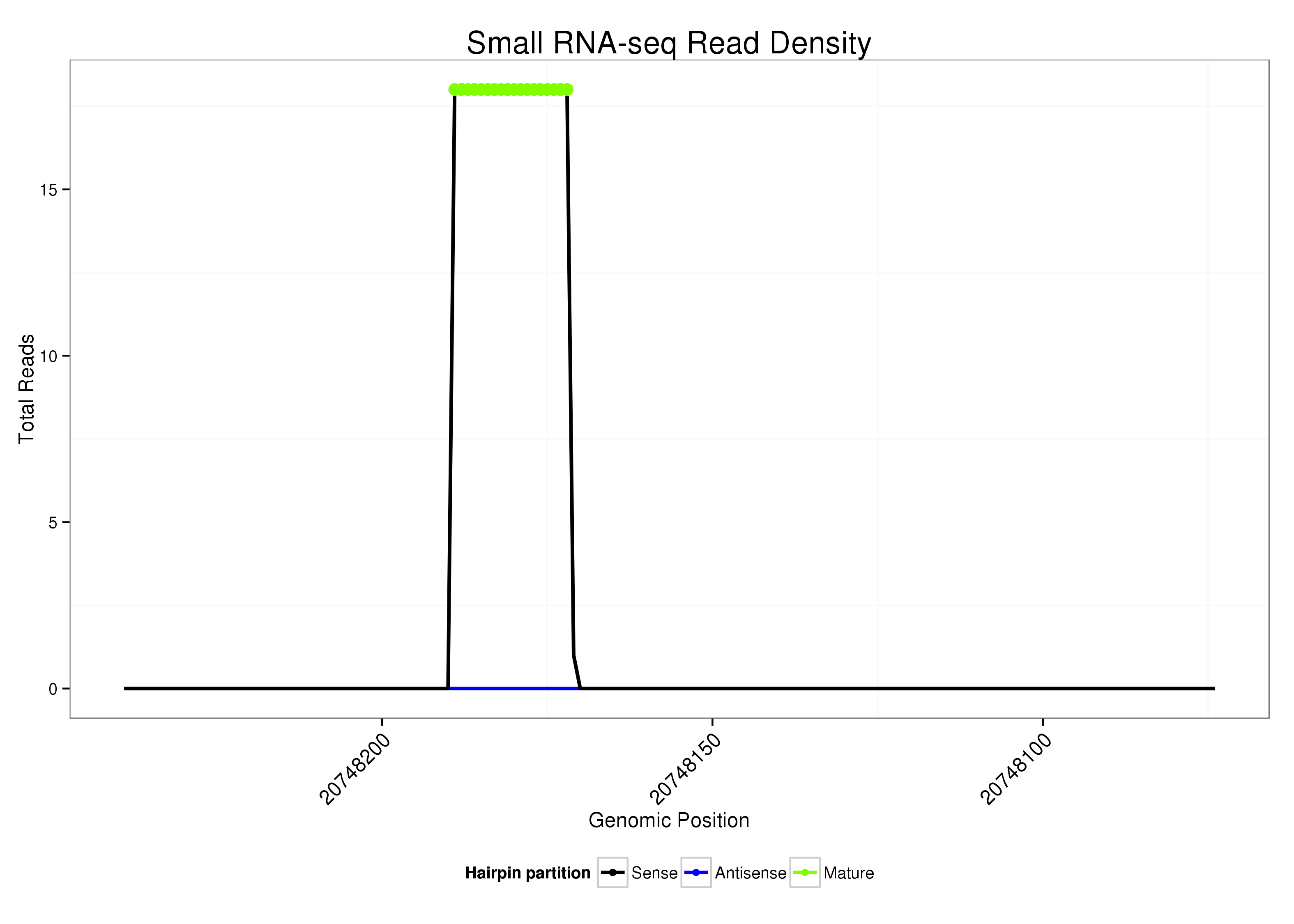
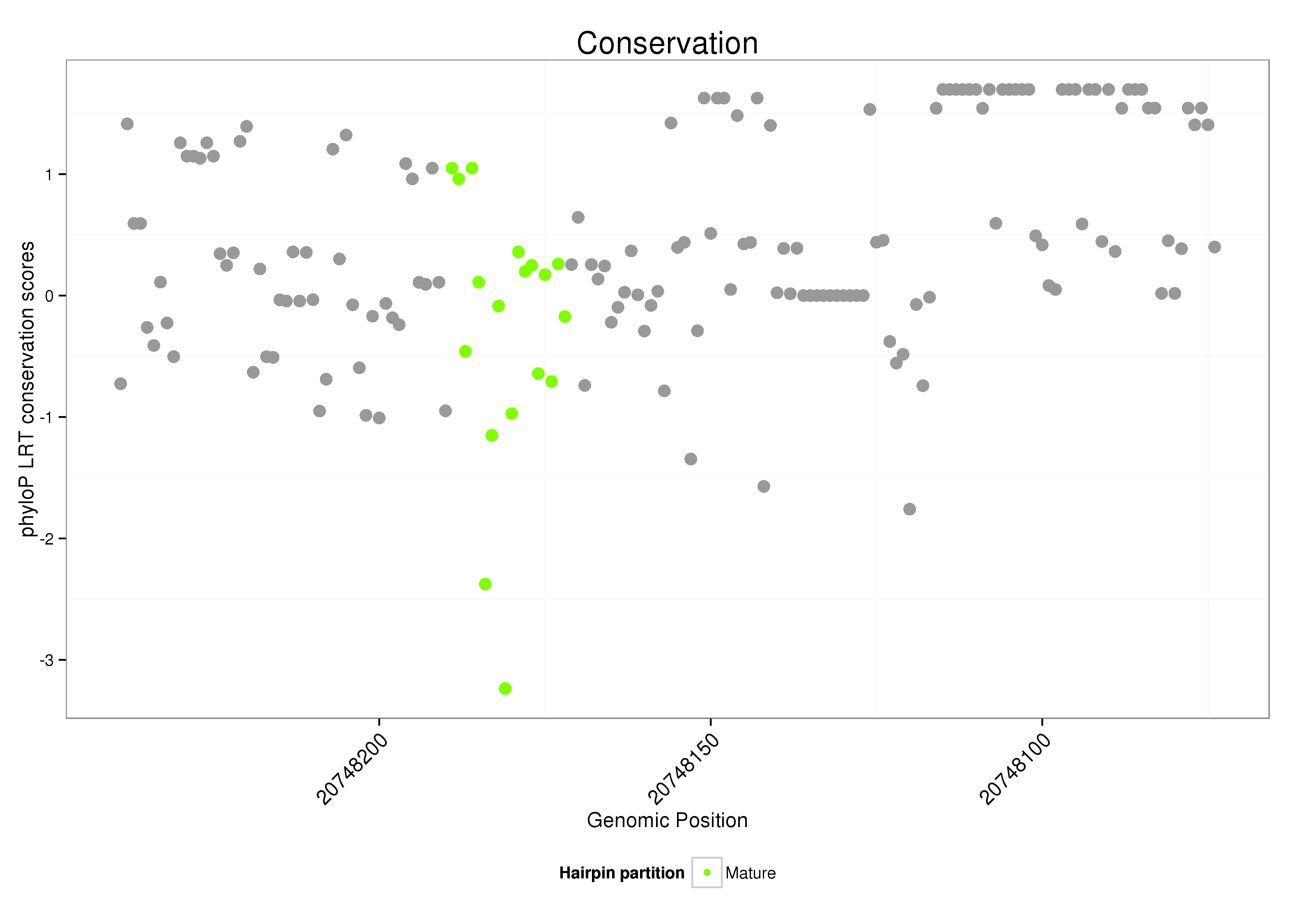
ID:dsi_261 |
Coordinate:3R:20748124-20748189 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -30.2 |
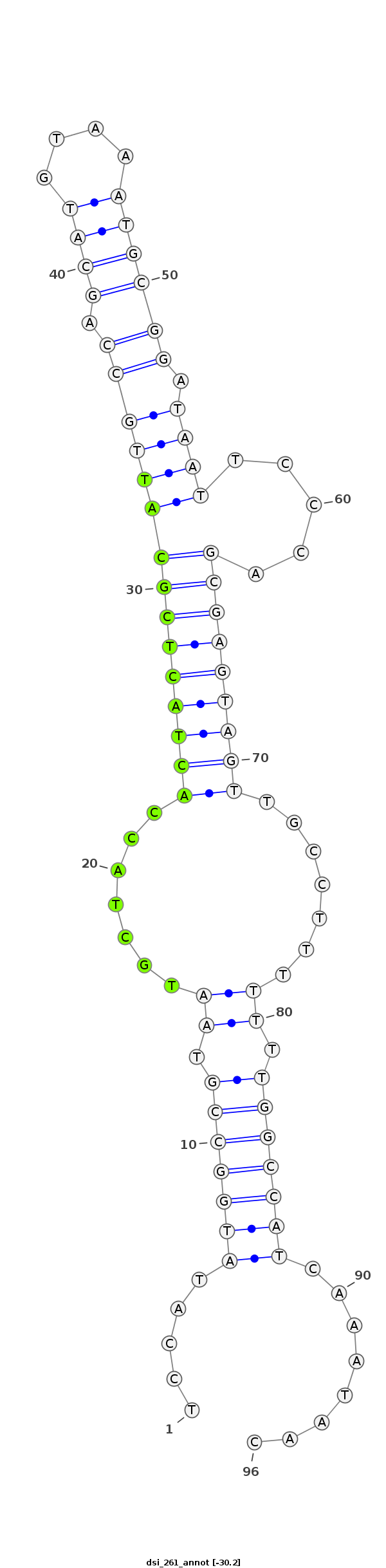 |
intron [Dsim\GD18236-in]
No Repeatable elements found
|
TGTAGAGTCTCGTAGAGAGTTGGCTATAATGCGATTCCATATGGCCGTAATGCTACCACTACTCGCATTGCCAGCATGTAAATGCGGATAATTCCCAGCGAGTAGTTGCCTTTTTTTGGCCATCAAATAACATATTTAGCCATATTATATCGATTTAGGGCACTGT
***********************************.....(((((((..........(((((((((((((((.((((....)))))).)))).....)))))))))..........)))))))........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O002 Head |
O001 Testis |
|---|---|---|---|---|---|---|---|
| ..................................................TGCTACCACTACTCGCAT.................................................................................................. | 18 | 0 | 1 | 17.00 | 17 | 16 | 1 |
| ....................................................................................................................................GATTTGGCCATACTATATCGAT............ | 22 | 3 | 1 | 2.00 | 2 | 0 | 0 |
| TGAAGAGTCTCGTCGAGAGAT................................................................................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 |
| ..................................................TGCTACCACTACTCGCATT................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................................................................ATATTATATCGATTTAGT....... | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 |
| .CTAGAGTCTTGTAGAG..................................................................................................................................................... | 16 | 2 | 12 | 0.17 | 2 | 0 | 0 |
| ..........................................................CTTCTCGCATTGCCAGCT.......................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 |
| .CTAGAGTCTTGTAGAGGG................................................................................................................................................... | 18 | 3 | 16 | 0.13 | 2 | 0 | 0 |
| .GTAGAGACTCGTTGAGAGG.................................................................................................................................................. | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 |
| .......................................................................................ATAATTCCCACCGAG................................................................ | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 |
| TGTAGAGTCTCGTAAAA..................................................................................................................................................... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 |
| .......................................................................................ATAATTCTCAGCGTGTA.............................................................. | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 |
| .CTAGAGTCTTGTAGAGG.................................................................................................................................................... | 17 | 3 | 20 | 0.05 | 1 | 0 | 0 |
| ......................................................................................................................................TTTAGCCATGTTATAGTGA............. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 |
|
ACAGTGCCCTAAATCGATATAATATGGCTAAATATGTTATTTGATGGCCAAAAAAAGGCAACTACTCGCTGGGAATTATCCGCATTTACATGCTGGCAATGCGAGTAGTGGTAGCATTACGGCCATATGGAATCGCATTATAGCCAACTCTCTACGAGACTCTACA
***********************************........(((((((..........(((((((((.....((((.((((((....)))).)))))))))))))))..........))))))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|---|---|---|---|---|
| ........................................................................................................GTAATGGTAGCATGACGG............................................ | 18 | 2 | 1 | 1.00 | 1 |
| ......................................ATTTGATGGACAAAAAAAGTC........................................................................................................... | 21 | 2 | 1 | 1.00 | 1 |
| ............................................................................................................................ATATGGAATCGGACTACAGC...................... | 20 | 3 | 8 | 0.63 | 5 |
| ..........................................................................................TGCTGGCAGTGGGAGAAGTGGT...................................................... | 22 | 3 | 3 | 0.33 | 1 |
| ................................................................................................................................................CAACTCTCTACGTGA....... | 15 | 1 | 6 | 0.17 | 1 |
| ........................................................................................................GTAATGGTAGCATGAC.............................................. | 16 | 2 | 20 | 0.05 | 1 |
| ............................................................................................................TCGCAGCATGACGGCCATA....................................... | 19 | 3 | 20 | 0.05 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | 3R:20748074-20748239 - | dsi_261 | TG----------------------TAGAGTCTCGTAGAGAGT--TGGCTATAATG--CGATTCCATA----TGGCCGTAATGCTACC----------------ACTACTCGCATT---------------------------GCCA-------------GCATGTA-AA--TGCGGATAATTCCCAGCGAGTAGTTGCCTTT---------TTTT-GGCCATCAAATAACATATTTAGCCATATTATATCGATTTAGGGCACTGT-------- |
| droSec2 | scaffold_33:179079-179245 - | TG----------------------TAGAGTCTCGTAGAGAGT--TGGCTATAATG--CGATTCCATA----TGGCCGTAGTGCTACC----------------ACTACTCGCATT---------------------------GCCA-------------GCATGTA-AA--TGTGGATAATGCCCAGCGAGTAGTTGCCTTTT--------TTTT-GGCCATCAAATAACACATTTAGCCATATTATATCGATTTAGGGCACTGT-------- | |
| dm3 | chr3R:20917934-20918108 - | TG----------------------TAGAGTCTCGTAGAGAGT--TGGCTATAATG--CGATTCCATA----TGGCCGTAATGCTACC----------------ACTACTCGCATT---------------------------GCCA-------------GCATGTA-AA--TGCGGATAATTCCCAGCGAGTTGTTGCCTTTTTTTTTTTGTTTT-GGCCATCAAATAACATATTTAGCCATATTATATCGATTTAGGGCACTGT-------- | |
| droEre2 | scaffold_4820:7132411-7132583 + | TG------------------------GAGTCTCGTAGAGAGT--GGGCTATAATG--CGATTCCAAA---GTGGCAGTAATGCTATC-----------TACCTACCACTCGCAATGAAATACTCGATGGGCATG----------CAGCATGTTAC----GTATGTA-AATGTGCGGATAATTCCCAC-----------------------------------CAAATAACATATTTAACCATATTATATCGATTTAGGGCACTGT-------- | |
| droYak3 | 3R:25578391-25578564 + | TGGAATTGGAGTTGGAGTTGAAGTTAGAGTCTCGTAGAGAGT--TGGCTATAATG--CGATTCCAAA---GTGGCAGTAATGCTACC----------------AGCACTCGCAATGAAATACTCGCT--------------------------------GTATGTA-AATGCGCGGATAATTCCCAT-----------------------------------CAAATAACATATTTAGCCATATTATATCGATTTAGGGCACTGT-------- | |
| droEug1 | scf7180000409692:481170-481305 + | AG-----------------------------------AGAGTTTCCAATATAACG--AGATCTTTTA----TAG------------G----------------ACTA--------------TACG-------------AT--GGTA-------------GCAAGTTTAA--TGCGTTTAATTCCCATCAAAT------------------GTTTCCGTGGACCAAATAACATATTTAGGAATATTATATCGATTTAAAACACTGTT------- | |
| droBia1 | scf7180000302136:1716222-1716421 - | AG------------------------AGGCGTCGTAGAGAGT--CGAACCCCATA--CGATTTCATATACATGGCAATGTTGTTTTA---AC--ACCTTGAGCTGCGCTCGAAATGAAATATTTTCCG-------------G--T-----GGTATGTATGCAAGTA-GA--GGCGTATAATTCCCACCAAAT------------------GTTTCAGGCGTCCAAATAACATATTTAGCCATATTATATCGATTTAAAACACTGCGTTCGAGT | |
| droTak1 | scf7180000415658:236539-236697 - | -------------------------------TCGTAGCGAGT--CGAATATAATA--CGATTTTA------GG---------------------ACCTTAAGCAACACTTGAAATGAAATATTCGATG-------------A--T-----GGTAA----GCAAGTA-AG--GGCGAATAATTCCCACCAAAT------------------ATTTCAAATTTTCAAATAACATATTTAGCAATATTATATCGATTTAAAACACTGC-------- | |
| droEle1 | scf7180000491212:239659-239824 - | AG------------------------AGATATCGTAGAAAGT--CGTATATAAAC--CGATTGTACG----TAGCAATAATGGTAGCAAAAAGGTCTTTATGCTTTACTTTTATTTTAATATTTA-------------------------------------------------ATATAATTCCTATCAACT------------------TTTCC-ACAGGCCAAATAACATATTTAGCAGTATTATATCGATTTAAAACACTGT-------- | |
| droRho1 | scf7180000779365:351666-351832 + | AG------------------------AGATATCGTAGAAAGT--CGAATATAAT------------------AGCAATAATGCTAAA---AT--ACCTTATGGTACATGATTGTTTAAATATTTG-------------ATAT--TA-------------ACCAGCT-AA--AGCGGATAATTCACATCGAAT------------------GTTTC-ACAGGCCAAATAACATATTTAGCAGTATTATATCGATTTAAAACACTGT-------- | |
| droFic1 | scf7180000454055:817777-817936 - | CG----------------------TAGAGCGTCG-AGAGTGT--CGAGTATAATA--GGATTT----------------------------------------AGCACTCGAAACGATATATG-TACATATACACAAAAT--GGTA-------------GTAAGTA-AG--AGCGTTTAATTCCCACCAAAT------------------GTTTC-GGCGGCCAAATAACATATTTAGCAATATCATATCGATTTAGAGCACTGT-------- | |
| droKik1 | scf7180000302388:996056-996224 + | TG------------------------AAGTGTCGTAGAGAGT--GGCCAGTGGTCAGTGGTCAGTCG----CGGCAATAGTGCTAAA---AAG-CCCTT--GCAATACTCGTAATTAAATATCTG-------------AAAG--TG-------------GCGAACA-TA--AGCGGATAATACCCATCACAT------------------GTTTT-AGAGGCCAAATAACATATTTAGCAGTATTATCTTGATT------------------- | |
| droAna3 | scaffold_13340:6780259-6780301 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAATAACATATTTAGCAATATTATCTTGATTTAAAATACTGT-------- | |
| droBip1 | scf7180000396708:2068183-2068266 + | CG----------------------------------------------------------------------------------------------------------------------------------------------------------------TGTG-TA--AGCGGCTGATTCCCACCAAAG------------------GTCTAAAGCGTCCAAATAACATATTTAGCAATATTATCTTGATTTAAAATACTGT-------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 12/06/2013 at 02:36 PM