| droSim3 |
2L:10397847-10397993 - |
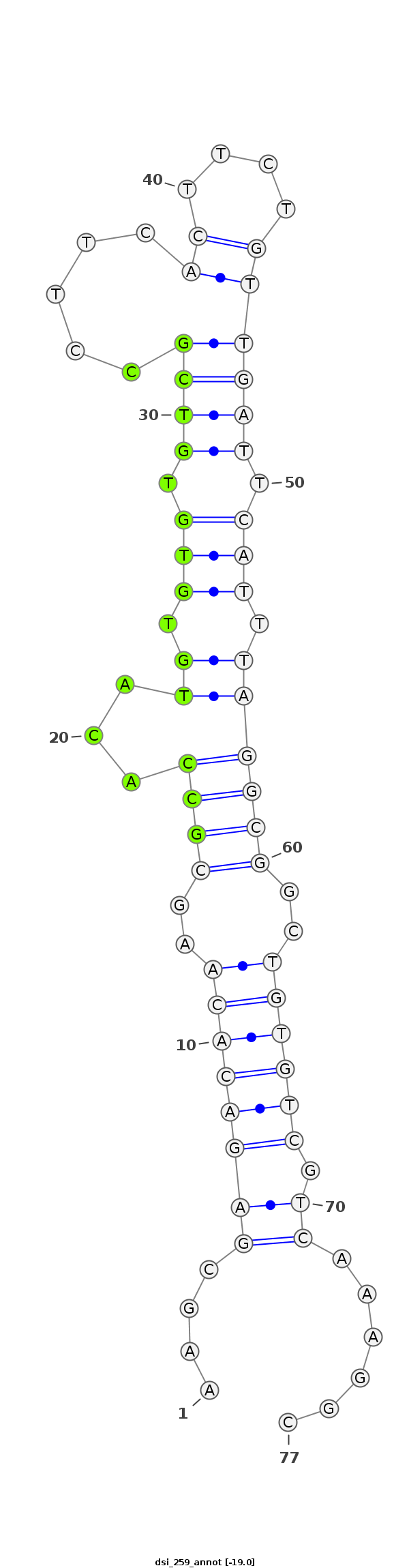
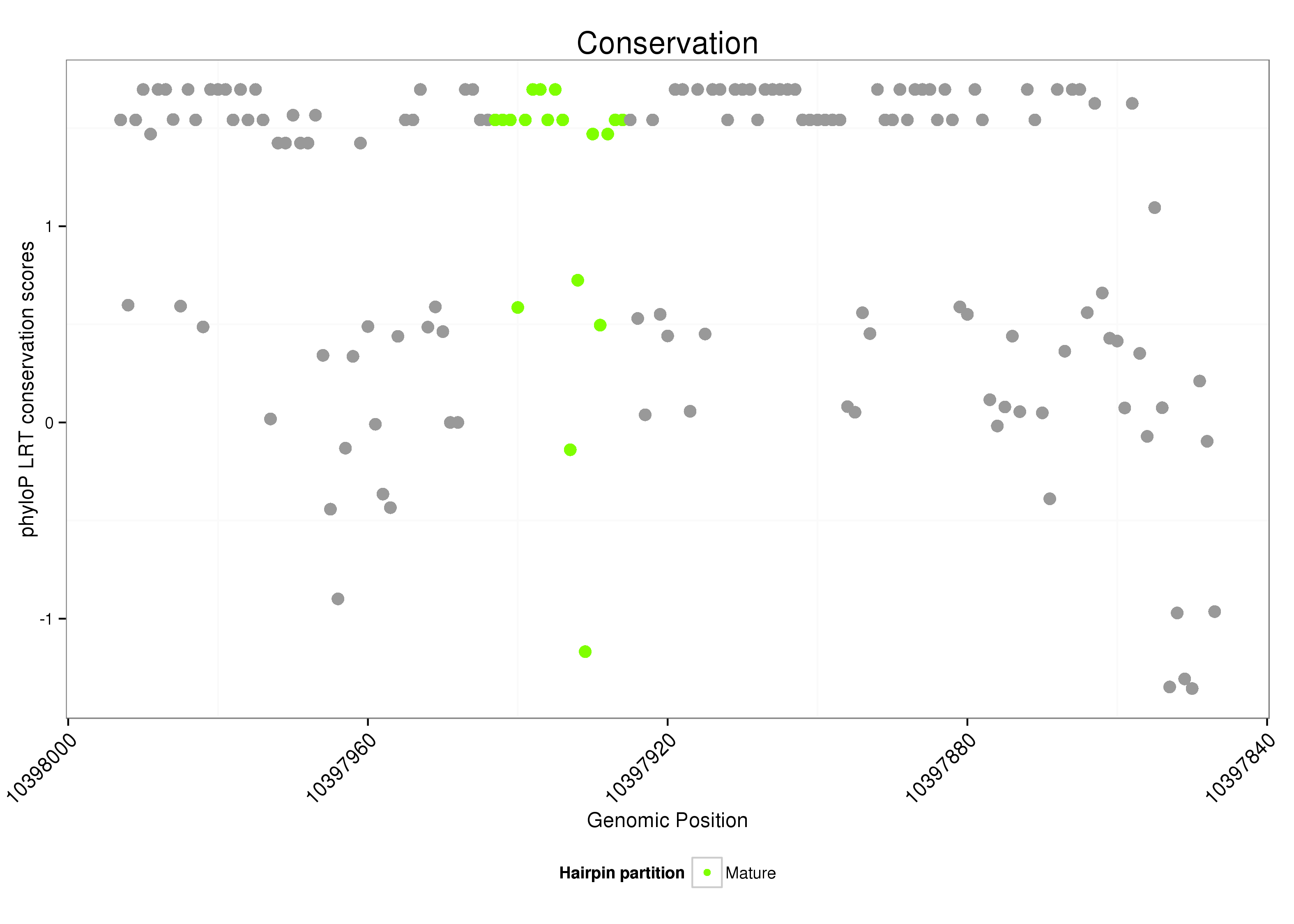
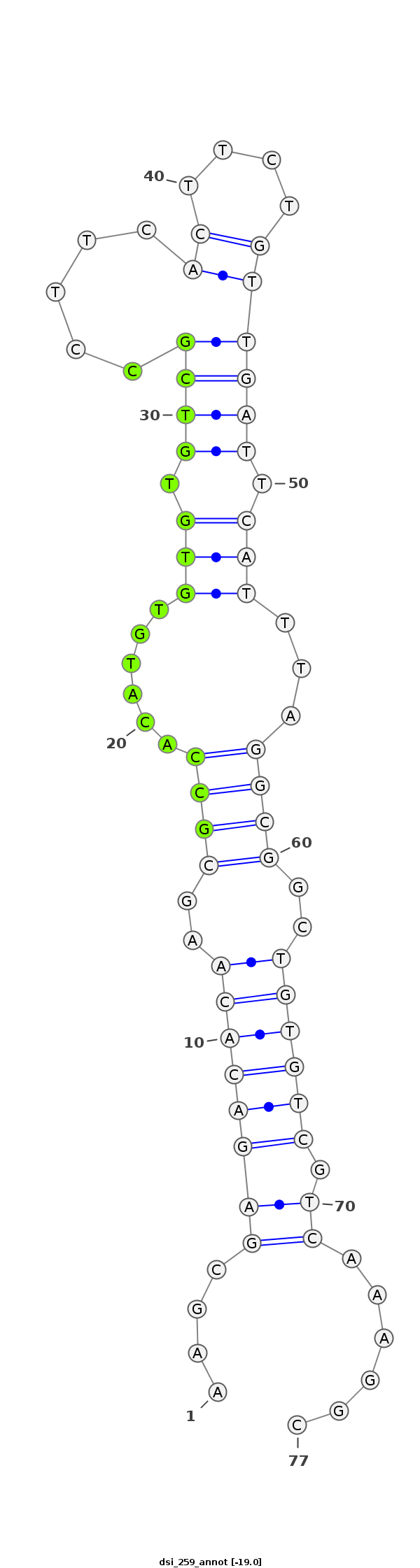
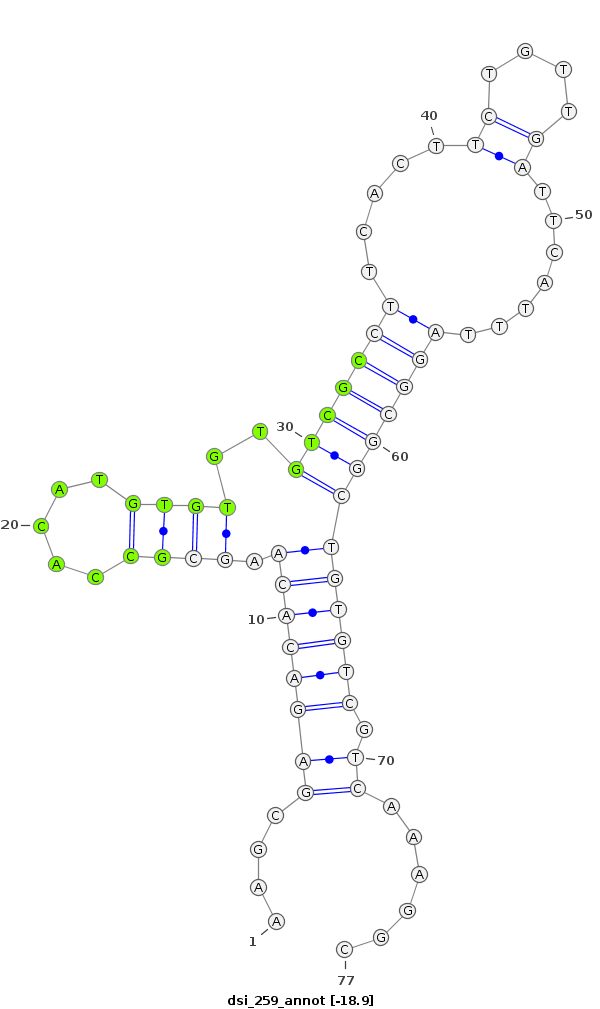
dsi_259 |
GACACTTTTTCCTTTGACAGTCGACGA-------------------------------CGTTGGAAAAGCGAGACACAAGCGCCACATGTGTG------TGTCGCCTTCACTTC---------------TGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGGCAAAG--------------AGTGTAGGCACTTTTTCCTTTT---------------TCG-C-------CATTA |
| droSec2 |
scaffold_3:6019315-6019461 - |
|
GACACTTTTTCCTTTGACAGTCGACGA-------------------------------CGTTGGAAAAGCGAGACACAAGCGCCCCATGTGTG------TGTCGCCTTCACTTC---------------TGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCAAAG--------------TGTGTAGGCACTTTTTCCCTTT---------------TCG-C-------CATTA |
| dm3 |
chr2L:10601102-10601247 - |
|
GACACTTTTTCCTTTGACAGTCGACGA-------------------------------CGTTGGAAAAGCGAGACATAAGCGCCACATGTGTG------TGTCGCCTTCACTTC---------------TGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCAAAG--------------AGTGTAGGCACTTTT-CCTTTT---------------TCG-C-------ATTTA |
| droEre2 |
scaffold_4929:14834035-14834183 + |
|
GGCACTTTGTCCTTTGACAGTCGACGA-------------------------------CGCTGGAAAAGCGAGAC--AAGCGCCACATGTG--------TGTCGCCTTCACTTC---------------TGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCAAAG--------------AGTGTAGGCACTTTT-CCTTTT----------------CGCATTCACGCATTTA |
| droYak3 |
2L:7003252-7003411 - |
|
GACACTTTTTCCTTTGACAGCCGACGACG--A--------------------------CGTTGGAAAAGCGAGAC--AAGCGCCACATGTG--------CGTCGCCTTCACTTCACTTCTGTTCTGTTCTGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCAAAG--------------AGTGTAGGCACTTTT-CCTTTT---------------TCG-C-------ATTTA |
| droEug1 |
scf7180000409554:4315960-4316105 + |
|
GACACTTTTTCCTTTGACAG------------------------------------------------GCGAGAC--AAGCGCCACATGTGTGTGTGTACGTCGCCTCCACTTC---------------TGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCAAAG--------------AGTGGAGGCACTTTT-CCTTTCATACATATACACTTTTCC-C--------A-TT |
| droBia1 |
scf7180000302422:8528300-8528435 - |
|
GACA-TTTTTCCTTTGACAGCCGACGA-------------------------------CGCTGGG-AGGCGAGAC--AAGCGCCACATGTG--------T---GCCTCCACTTC---------------TGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCAAAG--------------CGCGGAGGGACTTTT-CCTTTCTT---------------A-T--------TTTC |
| droTak1 |
scf7180000415160:247027-247165 - |
|
GACACTTTTTCTTTTGACAGCCGACGA-------------------------------CGTTGGAAGGGCGAGAC--AAGCGCCACATGTG--------CGTCGCCTCCACTTC---------------TGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCAAAG--------------AGTGGAGATACTTTT-CCTTTCTT---------------A-----------TTT |
| droEle1 |
scf7180000490971:290743-290884 + |
|
GACACTTTTTCCTTTGACAGCCGACGA-------------------------------CGTTGGAAAAGCGAAAC--AAGCGCCACATGTG--------CGTCGCCTCCACTTC---------------TGTTGATTCATTTAGGCGGCCATATCGTCAAAGTCAAAG--------------AGTGGAGGTACTTTT-CCTTTC---------------TGC-C-------CCCGT |
| droRho1 |
scf7180000780147:17626-17763 + |
|
GACACTTTTTCCTTTGACAGCCGACGACG--A--------------------------CGTTGGAAGGGCGAGAC--AAGCGCCACATGTG--------TGTCGCCTCCACTTC---------------TCTTGATTCATTTAGGCGGCTCTATCGTCAAAGTCAAAG--------------AGTGGAGGTACTTTT-CCTTTCCT------------------------------ |
| droFic1 |
scf7180000453838:862608-862749 - |
|
GACACTTTTTCCTTTGACAGCCGACGA-------------------------------CATTCGAGGGGCGAGGA--AAGCGCCACATGTG--------CGTCGCCTCCACTTT---------------TGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCGAAG--------------ACTGGAGTTACTTTT-CCTTTC---------------TCA-T-------TTTTC |
| droKik1 |
scf7180000302472:2730651-2730839 + |
|
GACACTT-TTCCTTTGACAGCCGACGACAAAAGGGAAGGAATGGAATGGAG---G---CAAAGGAAAATCGAGAC--AAGCGCCACATGTGAGT-TGC-TGCCGCCGCCACTTT--------TCTTTCTTGTTGATTCATTTAGGCGGCTGCGTCGTCAAAGTCAAAGTCAATGC------TATTGGAGGTATTTTT-CTTTATT-----------TTTTCT----------CTCC |
| droAna3 |
scaffold_12916:14441041-14441202 - |
|
GACACTTTTTCCTTTGACAGCCGACGAA--AAGGAAAAAAAAAGAAAGCCG--------GCAGGAGAAGCGAGAC--AAGCGCCACATGTG--------CGTCGCCTTCGATTC--------------TTGTTGATTCATTTAGGCGGCTGTGTCGTCAAAGTCAGAGTCAGAGTCAGCCAAAGTGAAGGCATTTA---------------------------------------- |
| droBip1 |
scf7180000396572:2533905-2534072 + |
|
GACACTTTTTCCTTTGACAGCCGACGAA--AAGGAAAAA--AAGACAGACAGAAGCCTTCTCGGAGAAGCGAGAC--AAGCGCCACATGTG--------CGTCGCCTCCGATTC--------------TTGTTGATTCATTTAGGCGGCAGTGTCGTCAAAGTCAGAGTCAGA------CAAAGCTAAGGCATTTATTTCCT---------------------------------- |