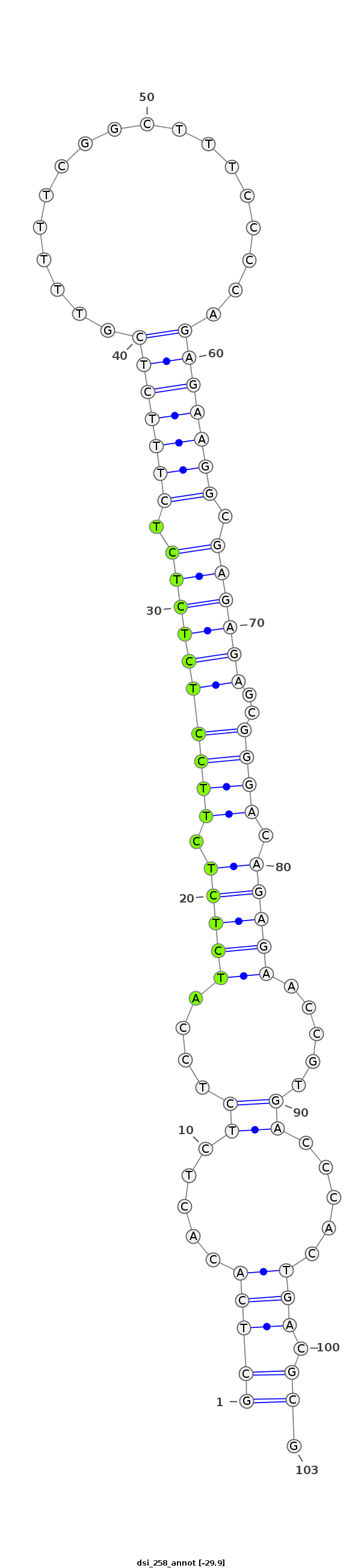
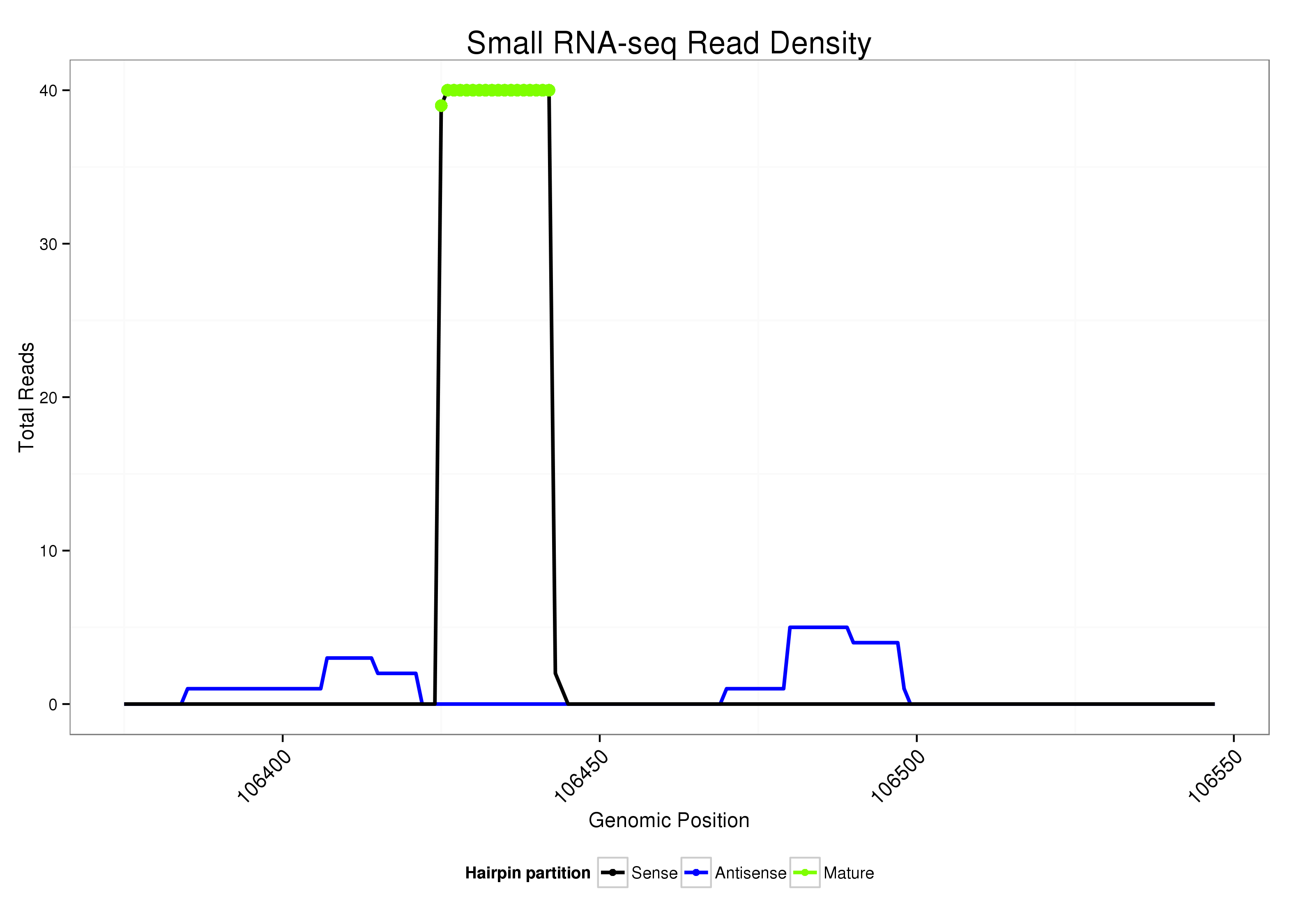
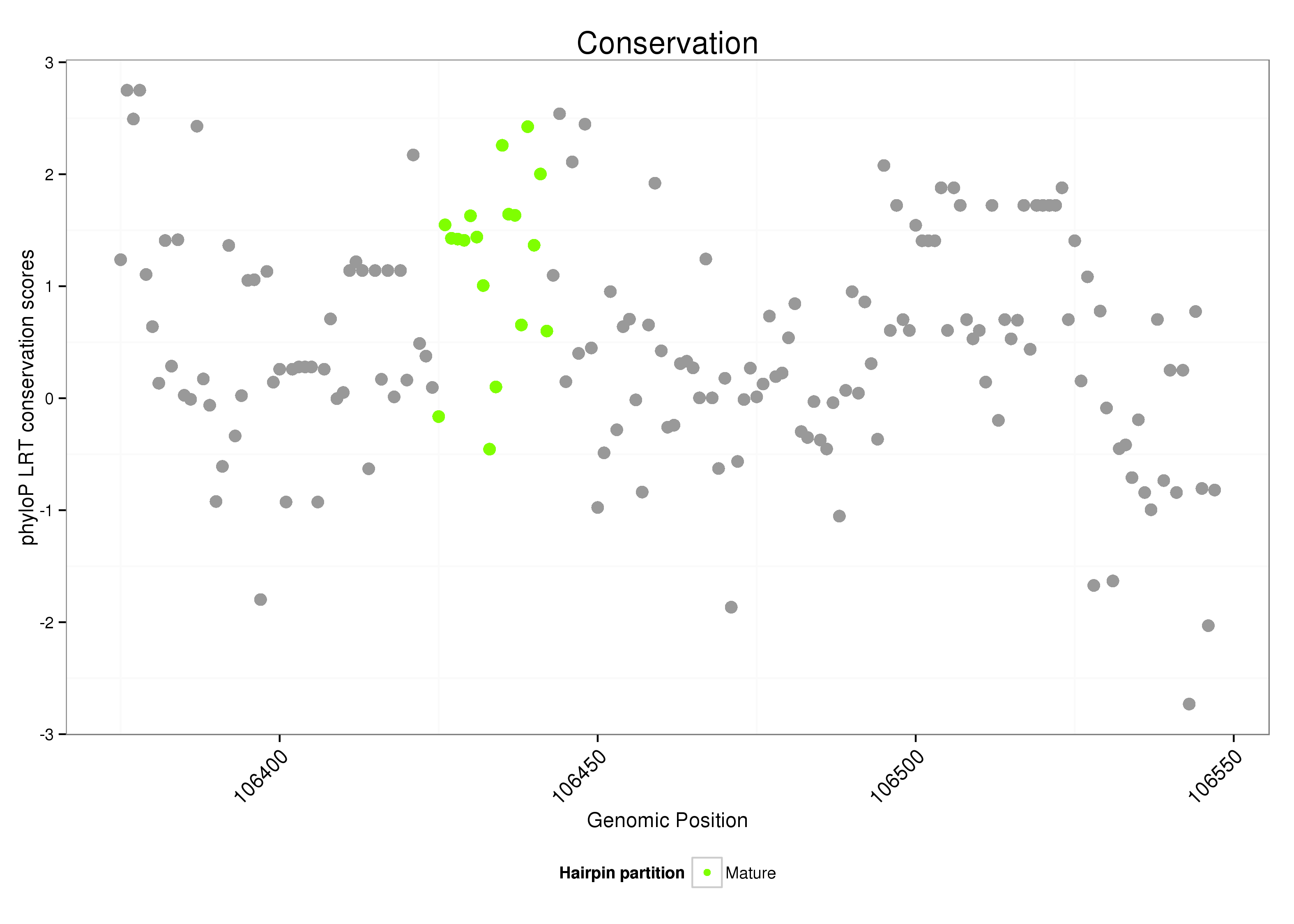
ID:dsi_258 |
Coordinate:chrX_Mrandom_022:106425-106497 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -29.9 | -29.6 | -29.6 |
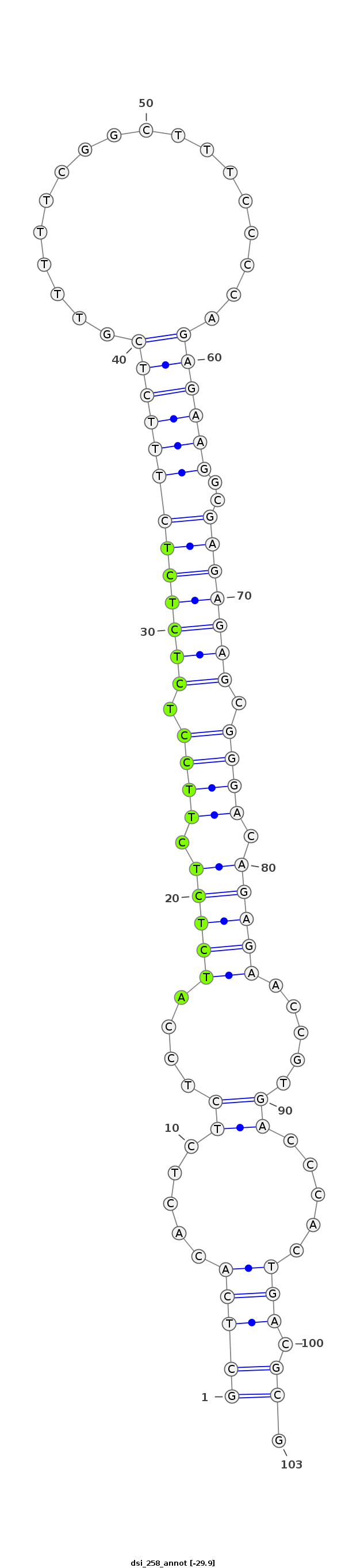 |
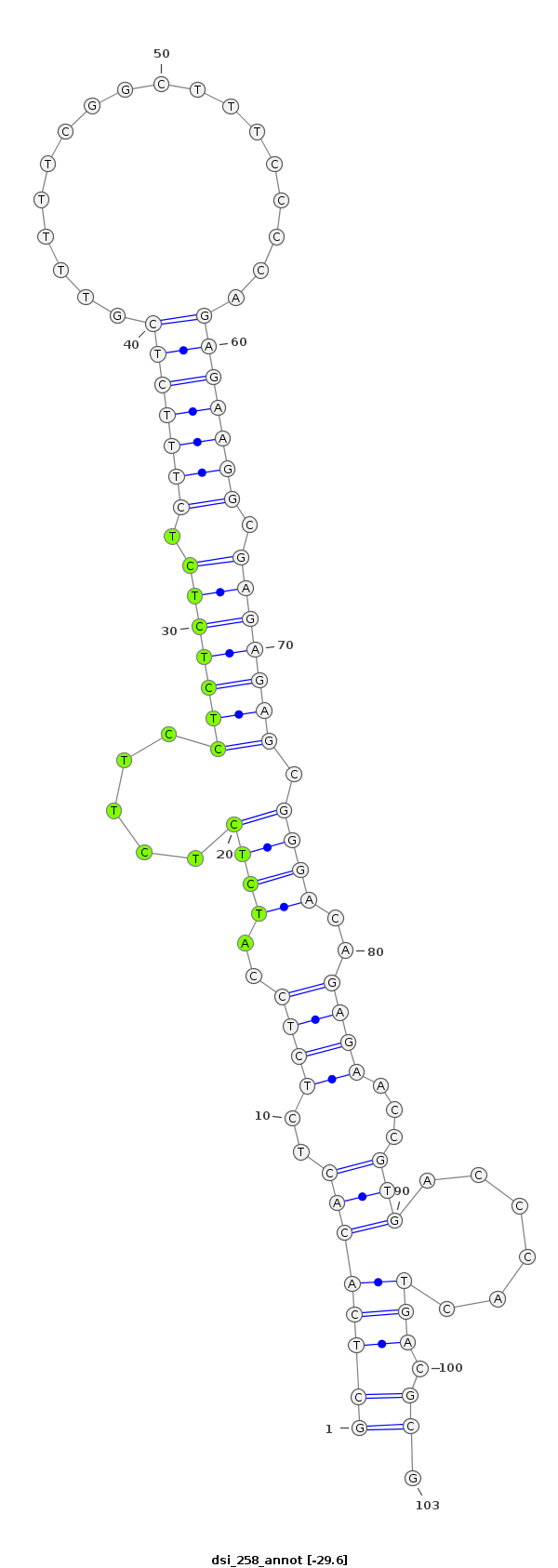 |
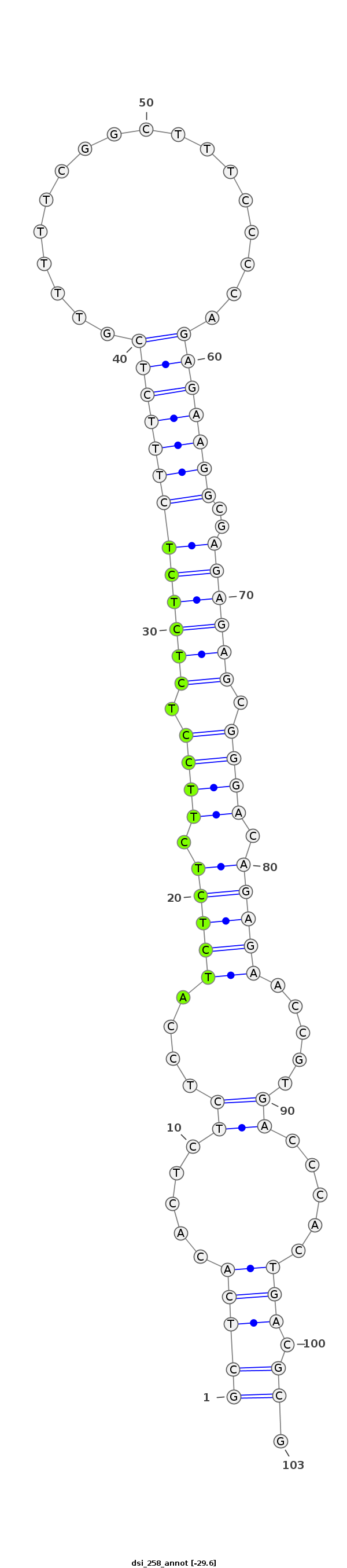 |
intergenic
| Name | Class | Family |
| CT-rich | Low_complexity | Low_complexity |
|
CTCTCCATTTCCCCCATTGCCCCCTCACTTTCCCCGCTCACACTCTCTCCATCTCTCTTCCTCTCTCTCTTTCTCGTTTTTCGGCTTTCCCCAGAGAAGGCGAGAGAGCGGGACAGAGAACCGTGACCCACTGACGCGTTGACCGGCCTACTGTGAATGTGCATATGTGTGTG
***********************************(((((.....((....(((((.((((((((((.(((((((..................))))))).))))))..)))).))))).....)).....))).)).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O002 Head |
O001 Testis |
|---|---|---|---|---|---|---|---|
| ..................................................ATCTCTCTTCCTCTCTCT......................................................................................................... | 18 | 0 | 1 | 38.00 | 38 | 37 | 1 |
| .............................................................................................................................AACCACTGACGCGTTG................................ | 16 | 1 | 1 | 2.00 | 2 | 0 | 0 |
| ...................................................TCTCTCTTCCTCTCTCTC........................................................................................................ | 18 | 0 | 3 | 1.00 | 3 | 3 | 0 |
| ..................................................ATCTCTCTTCCTCTCTCTCT....................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ..........................................................................................................................ATGGCCCACTGACGCGTT................................. | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 |
| ..........................................................................................................................ATGGCCCACTGACGCGT.................................. | 17 | 2 | 8 | 0.25 | 2 | 0 | 0 |
| .................................................................................................................................ACTGACGCGTTGAAC............................. | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 |
| .................................................................................................................................ACTGACGCGTTGCGCG............................ | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 |
| .........................................................................TCGTTTTTCGGCTTTA.................................................................................... | 16 | 1 | 13 | 0.08 | 1 | 0 | 0 |
| ..............................................................................ATTCGGCTTTCCCCATAGT............................................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 |
|
CACACACATATGCACATTCACAGTAGGCCGGTCAACGCGTCAGTGGGTCACGGTTCTCTGTCCCGCTCTCTCGCCTTCTCTGGGGAAAGCCGAAAAACGAGAAAGAGAGAGAGGAAGAGAGATGGAGAGAGTGTGAGCGGGGAAAGTGAGGGGGCAATGGGGGAAATGGAGAG
***********************************.((.(((.....((.....(((((.((((..((((((.(((((((..................))))))).)))))))))).)))))....)).....)))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O002 Head |
O001 Testis |
SRR618934 Ovary |
SRR553485 Ovary |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................AGAGAGAGGAAGAGAGAT.................................................. | 18 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 |
| ................................CAACGCGTCAGTGGA.............................................................................................................................. | 15 | 1 | 5 | 2.20 | 11 | 0 | 0 | 11 | 0 |
| ................................CAACGCGTCAGTGGG.............................................................................................................................. | 15 | 0 | 2 | 2.00 | 4 | 0 | 0 | 4 | 0 |
| .................................AACGCGTCAGTGGGCCA........................................................................................................................... | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .........................................................................................................AGAGAGAGGAAGAGAGATG................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................CAACGCGTCAGTGGATGACG......................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..........TGCACATTCACAGTAGGCCGGTCAACGCGT..................................................................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ................................CAACACGTCAGTGGGT............................................................................................................................. | 16 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................AACGAGAAAGAGAGAGAGGA.......................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..........................ACCGTGCAACGCGTCAGTGGG.............................................................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ................................CAACGCGTCAGTGGGCCA........................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...............................TCAACGCGTCAGTGGGC............................................................................................................................. | 17 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ................................CAACGCGTCAGTGGGA............................................................................................................................. | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 |
| ................................CAACGCGTCAGTGGGCTA........................................................................................................................... | 18 | 2 | 6 | 0.33 | 2 | 0 | 0 | 2 | 0 |
| ................................CCACGCGTCAGTGGG.............................................................................................................................. | 15 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 |
| ...............................GCAACGCGTCAGTGGGT............................................................................................................................. | 17 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 |
| ...............................GCAACGCGTCAGTGGGTG............................................................................................................................ | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ............................CGCGCAACGCGTCAGTGGG.............................................................................................................................. | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ......................GTAGTCCGGTCATCGCG...................................................................................................................................... | 17 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 |
| ................................CAACGCGTCAGTGGGGT............................................................................................................................ | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
| ................................CAACGCGTCAGTGGCGC............................................................................................................................ | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................TGGAGAGCGTGTGAG.................................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ................................CAACGCGTCAGTGGGTTGA.......................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ..................................ACGCGTCAGTGGGCTA........................................................................................................................... | 16 | 2 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ........................................CTGTGGGTCGCGGTTC..................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 |
| ............................CGCGCAACGCGTCAGT................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................TGCAATGGAGGAAATGGACA. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 |
| ......................GTAGTCCGGTCATCGC....................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | chrX_Mrandom_022:106375-106547 + | dsi_258 | CTCTC----CATTTCCCCCATTGCC-CC------CT----------------------------------------CACTTTCCCCGCTCACA---CTCTC---TCCATCTCT--CTT------------------CCTCT-CT--C------------T--CTTTCT--C--GTTTTTCGGC----TTTC-CC-C----------AGA-------------------------------------GAAGGCGA--G------------AGAGCGGGA---------C---AGAGAACCGTGACCCACTGACGCGTTGACCGGCCTACTG-----------------TGAATGTG--CAT--ATGTGTGTG---------------- |
| droSec2 | scaffold_15:851712-851884 + | CTCTC----CATTTTTCCCATTGCC-CC------CT----------------------------------------CACTTTCCCCGCTCACA---CTCTC---TCCATCTCT--CTT------------------CC--T-CT--C------------T--CTTTCT--C--GTTTTTCGGC----TTTC-CC-C----------AGA-------------------------------------GATGGAGA--G------------AGAGCGGGA---------C---AGAGAACCGTGACCCACTGACGCGTTGACCGGCCTACTG-----------------TGAATGTG--CATATGTGTGTGTG---------------- | |
| dm3 | chrX:10141542-10141728 + | CTCTC----CATTTTACCCCATTACCCC------CT----------------------------------------CACTTTCCCCGCTCACA---CTCTCTCCTCCATCTGT--CTGCCTCT----CT----CTCTCTCT-CT--C------------T--CTTTCT--C--GTTTTTCGGC----TTTG-CC-C----------AGA-------------------------------------GTGGGAGA--G------------AGAGCGGGA---------C---AGAGAACCGTGACCCACTGACGCGTTGACCGGCCTACTG-----------------TGAATGTG--CAT--ATGTGTGTG---------------- | |
| droEre2 | scaffold_4690:4979442-4979605 - | CTCTC----CATTTTCCCCATTTTCCCC------------------ATTTCC----CTCACTCTC-----------------------------------C---TCCATCTCT--C--------------------CCTCT-CT--C------------T--CTTTCT--C--GTTTTTCGGC----TTTC-CC-C----------AGC-------------------------------------GTGGAAGA--G------------AGAGCGGGA---------C---AGCGAACCGTGACCCACTGACGCGTTGACCGGCCTACTG-----------------GGAATGTT--CAT--ATGTATGCG---------------- | |
| droYak3 | X:18722392-18722579 + | CTCTCTCTCCATTTTCCCCCGTTTCCC--------T----------------------------------------CGCTTTTCCCGCTCCCA---CTCTC---GCCATCTCT--CTCCCTTC----CT----CTCTCTCT-CT--C------------T--CTTTCT--C--GTTTTTCGGC----TTTC-CC-C----------AGA-------------------------------------GAAGAAGAGAG------------AGAGCGGGA---------C---AGAGAACCGTGACCCACTGACGCGTTGACCGGCCTACTG-----------------TGAGTGTG--CAC--ATGTATGCG---------------- | |
| droEug1 | scf7180000409509:726005-726215 - | CTCTC----CATTTTCCCCACTCTCTCT------------------CTCTCT----GTTGCG-----------------------------------------------------------------------------CT-CT--C------------T--CTTTCT--C--GTTTTTCGGC----TTTCCCCC-GGTGACCCCA-AAGGAAGACAGAACGAGAAAGATATATAGAGC-GATAGAGAGAGAGA--G------------AGAACGGGA---------T---GGAGAACCGTGACCCACTGACGCGTTGACCGGCCTACGGATA-TGGTTACGGCCGTAGAATATGCGGAT--ATATGTATG---------------- | |
| droBia1 | scf7180000302187:1240070-1240245 - | CTCTC----CATTTCCCCATTTCCCCGC------------------GCTTTC----CTTTCTCTCCCTCTCT-------------------------------------------------------------------------------------------------------TTTTCGGC----TTTC-CCC-GGTGACCCCG-GG----GACAGA--------------GATAGC-GATA-----GGAGG--GCGGGCGCCGGCTAGAGCGGGA---------C---GGAGAGCCGTGACCCACTGACGCGCTGACCGGCCTACGGGTA------------------CGCG--GCT--ATACATGGGA--------------- | |
| droTak1 | scf7180000415872:830071-830201 + | CTCTC----CATTTTC------------------------------------------------------------------------------------------------T--CTC------------------CCTCT-CT--C------------T--CT-----------TTTTCGGCTCTTTTTC-CCC-GGTGACCCC-AGA-------------------------------------GCGGGA----G------------AGAGGGAGA---------G---GGAGAACCGTGACCCACTGACGCGTTGACCGGCCTACGGATA--------------------CG--ATT--ATATGGATA---------------- | |
| droEle1 | scf7180000491023:75664-75852 - | CTCTC----CAATTTTCCCACTCTCTCT------------------CTCTCT----CTCTCTCTCCCTCTCT----------------------------C---TGT--------------------CTCCATCTATC---TCGC----TCCCGCTGCCGCGTT--TTCCCA-CATTTTCCGC----TTTC-CCC-GGTGACCCCG----------------------------------------------GA--G------------AGAAAGGGA---------C--GGGAGAGCCGTGACCCACTGACGCGCTGACCGGCCTACGGATA-C-------------GGTTACG--ATT--ACGGCTGCG---------------- | |
| droRho1 | scf7180000779985:271298-271478 + | CTCTC----CAATTTCCCCACTTTCCCA------------------CTCTCT----CTCTCT-------------------------------------------------CT--CTCCCTTTCTCTCTGTCGCTTTC---TCTC----TCCC------T----------------------C----TGTC-CCC-GGTGGCCCCG----------------------------------------GTGGGA----G------------AGGGGGGAG---------G---GGAGAGCCGTGACCCACTGACGCGTTGACCGGCCTACGGATA-CGGTTACGGCTG-CGGGCAAGCGGAT--ATATGTGGA---------------- | |
| droFic1 | scf7180000454073:3074913-3075106 + | CTCTC----CATTTTCCCATTTTCCACACACACACACACTCTCTCTCTCTC--------------GCTCTCTCTGT------------------------C---TCTATC--------------------GGTGTCTCTCC-CT--C------------T--CTCTCT--T--ATTTTTCGGC----TTTC-CCC-TTTGACCCCG--------------------------------------------AGGA--G------------CGAGCGGGA---------T---GGAGAGCCGTGACCCACTGACGTGTTGACCGGCCTACGGATTTTGGTTACGGCTA-C--------------GAATATGCG---------------- | |
| droKik1 | scf7180000302476:777076-777193 - | CTCTC----TCTCTCTCTCTCTCTCTCT------------------CTCCTC----CTCTCTCTCTCTCTCT----------------------------C---TTTATCTCT--C------------------TTTC---TCTCTC------------T--CTCTCT--C--GATCTTGTGC----GGCA-TC--------------T-------------------------------------GCAAGTGA--A------------AGAATTGAA---------A---AGAGAAC----------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13248:3611201-3611342 - | CTCTC----TATTTTTCCCACACACACA------------------CACACACACA----------------------------CCACTCACACCACACTC---TCCTTCTCT-----------------------------CT--C------------A--CTCTCT--C--G----------------------AG-----------------------------------TGGG-CGGA----TAAGGAG---------------------GAG-GAGGCGTGGCTAGGGGGGGCCGTGACCCACTGACGCGCTGACCGGCCTAC----------------------------------------------------------- | |
| droBip1 | scf7180000392902:48646-48778 + | CTCTC----TATTTTTCCTACACACAC----------------------------------------------------------CACTCACACCACACTC---GCCGTCTCT--CTT------------------ACTCT-CT--C------------A------------------------------------AGTAG--------------------------------TGGG-CGGA----GGAGGAT---------------------GAG-GAGGCGTGGCTAGGGGGGGCCGTGACCCACTGACGCGCTGACCGGCCTAC----------------------------------------------------------- | |
| dp5 | XL_group1e:8980562-8980652 - | CTCTC----TCTCTCTCTCTCTCTCCCT------------------CTCTCT----CTCTCTCTCTCTCTCT----------------------------C---TCT--------------------CTCTCTCTCTC---TCTC----TC--------T--CTCTCT--C--TCTGTTCCTC----TTGC-CC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_5:6179016-6179127 - | CTCTC----TATCTCTCTCCCTCTCACACTCTCTCTCTCTCTCTCTCTCTC--------------TCTCTCT----------------------------C---TCAC----------------------------TC---TCTC----TCTCTCTCTCTCTCT--CTCTC----------------TCTCTC----------------------------------------------------------------------------------------------------------------------------------------------------------------------T--CTCTCTCTCGCTAGAACGAG----- | |
| droWil2 | scf2_1100000004909:6783828-6783958 - | TTCTG----CATTTTCCTTCTTCTGTCTCGCTCTCTCACTCTCTCTCTCTC--------------TCTCTCT----------------------------C---TCTC------------TCT----CT----CTCTCTCT-CT--C------------TTTCT--CTCTC----------------TTTCTCTC-TTTATCCTCG-TT-------------------------------------GAATGTCA--A------------AGAACACAA---------T---AGAAAAC----------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_13047:17104171-17104305 - | CTCTC----TCTCCCTCACTTTGTC------------------------------------------------------------GGCTCTCG---CTCAC---TCTCTCTCTCTC--------------------TCTCT-CT--C------------T--CTCTGT--C--GTT----GGC----TTAT-TG-C----------AGA-------------------------------------CAAG----------------------GCCTTC---------C---AAGGCACTGCA----ATTGGAATGTCTTCGGGCCTG--------------------------TC--GTT--GTGTGTGTG---------------- | |
| droMoj3 | scaffold_6540:8169612-8169727 - | CTCTC----TCTCTCTCTCTCTCTCTCT------------------CTCTCT----CTCTCTCTCTCTCTCT----------------------------C---TCT--------------------C--------TC---TCTCTCTCTCTC------T--CTGTCT--T--ATAAGAGCGC----CACT-TC-T----------AGA-------------------------------------GAGGGAGAGAG------------GCAGCGGCG---------C---AG---------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15116:804997-805101 + | CTCTC----TCTCTCTCTCTCTCTCTCT------------------CTCTCT----CTCTCTCTCTCTCTCT----------------------------C---TCT--------------------CTCTCTCTCTC---TCTC----T------------CTCTCT--CTCTCTC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--CTCTCTCTCGATAAAGGAAGAACGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 12/06/2013 at 02:36 PM