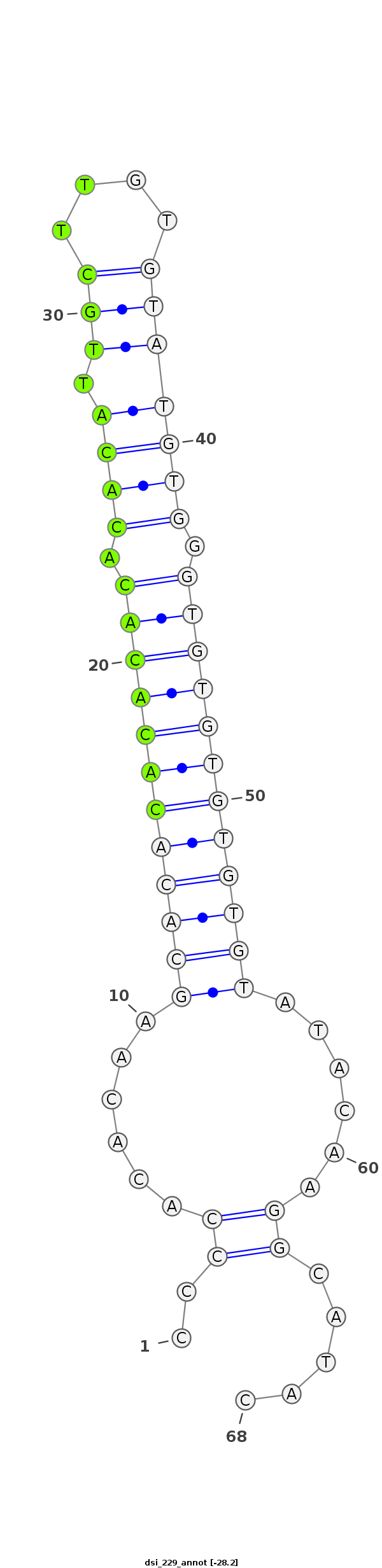
| droSim3 |
2L:1683093-1683230 + |
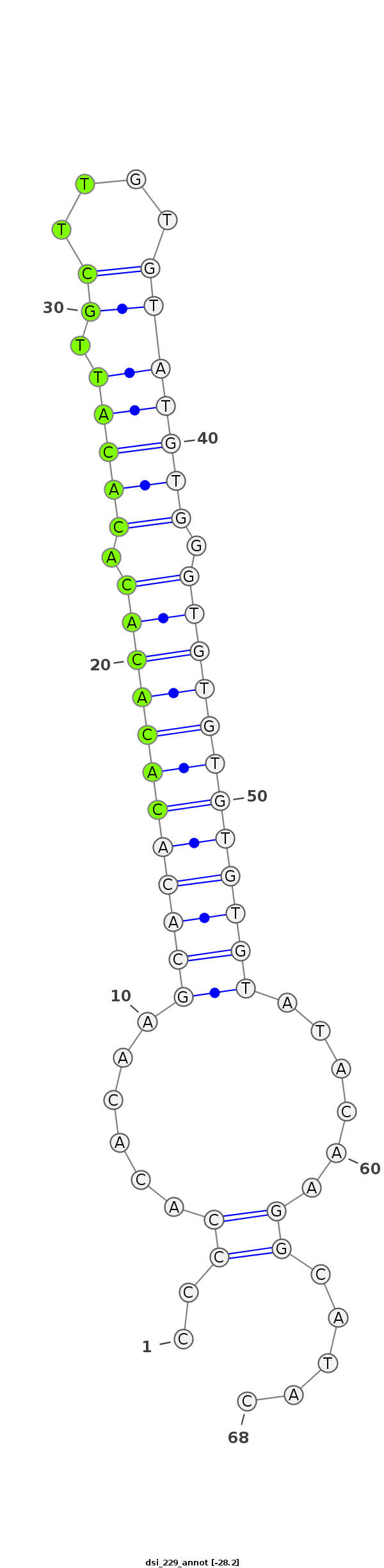
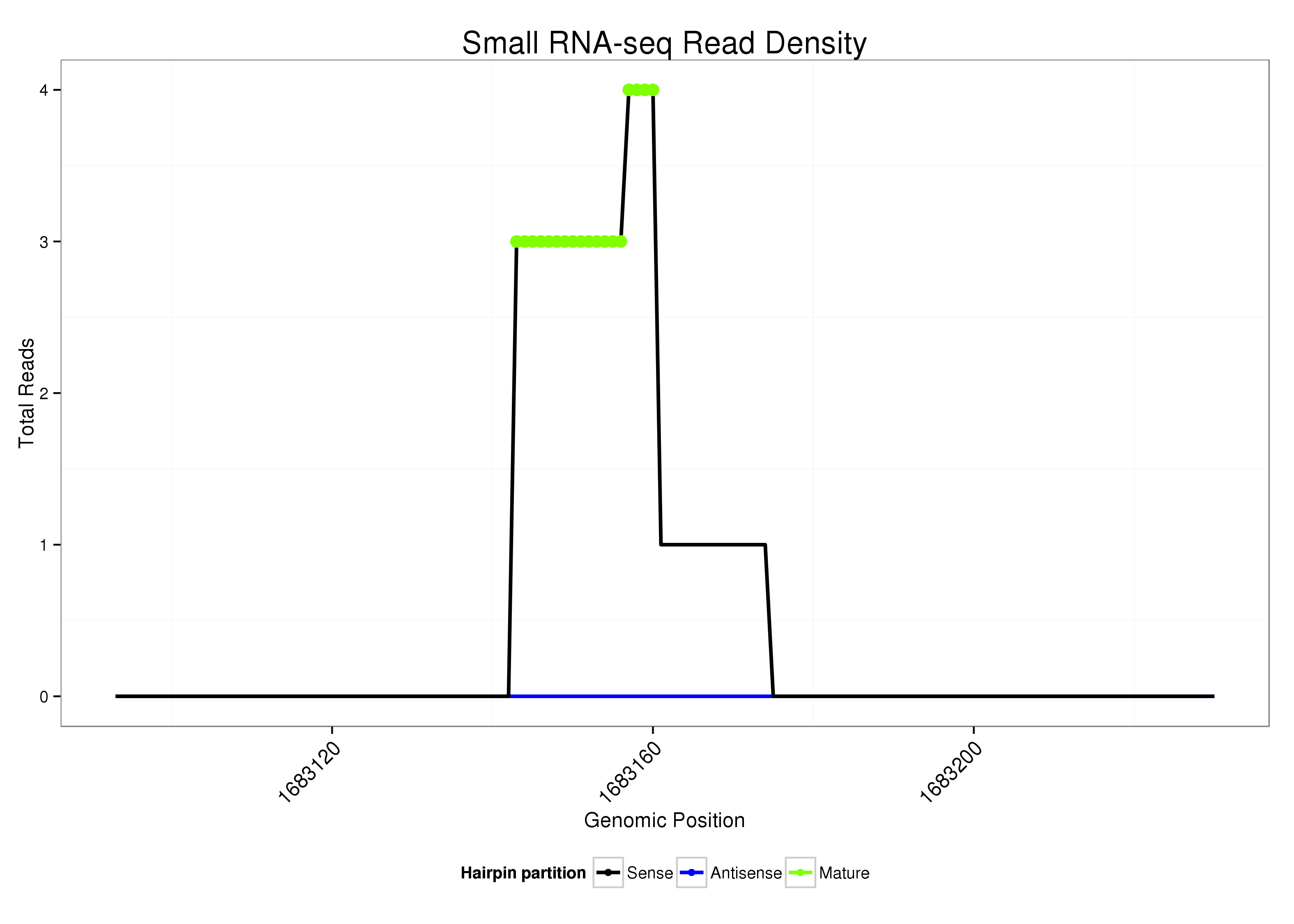
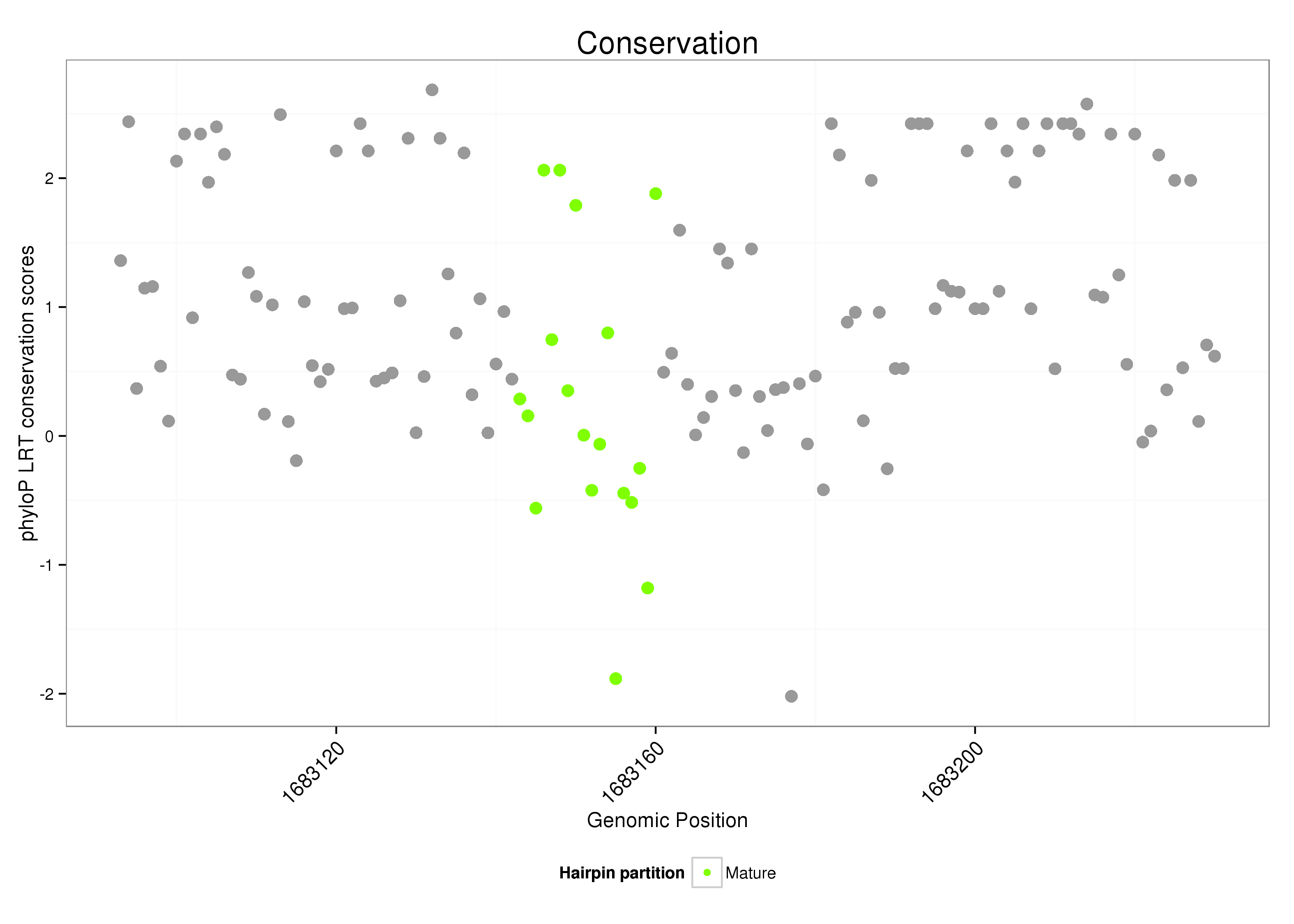
dsi_229 |
ACCAATGCAAACAC----------CCA------------------------------------------CACAAGCAC-A------------------------------CGCACGCGCCCCACA------CAAGCACAC--ACACACAC--ACATTGCTTGTG--------TAT-------------------GTGGGTGTGTGT-----GTG--TATAC-A-AGGCATACGAACGCAAGCTGCAAAAGTGCGTTCGG---AAACAGAA |
| droSec2 |
scaffold_14:1591786-1591921 + |
|
ACCAATGCAAACAC----------CCA------------------------------------------CACAAGCAC-A------------------------------CGCACGCGCCCCACA------CAAGCACAC--ACACACAC----ATTGTTTGTG--------TAT-------------------GTGTGTGTGTGT-----GTG--TATAC-A-AGGCATACGAACGCAAGCTGCAAAAGTGCGTTCGG---AAACAGAA |
| dm3 |
chr2L:1649067-1649196 + |
|
ACCAATGCAAACAC----------CCA------------------------------------------CACAACCAC-A------------------------------CGCACGCGCCCCACA------CAAGCACAC--ACACACAGAC------GTTGTGTGTAT--GTG----------------------------------CGTGTG--TATAC-A-AGGCATACGAACGCAAGCTGCAAAAGTGCGTTCGG---AAACAGAA |
| droEre2 |
scaffold_4929:1698469-1698600 + |
|
ACCAATGCAAACAC----------CCA------------------------------------------CACAACCAC-A------------------------------CGCACGCGCCCCACA------CAAGCACAC--ACACACACACGCACTCATTGTG----T--GTG----------------------------------GGTGTG--TATAC-A-AGGCATACAAACGCAAGCTGCAAAAGTGCGTTCGA---AAACAGAA |
| droYak3 |
2L:1624156-1624289 + |
|
ACCAATGCAAACAC----------CCA------------------------------------------CACAACCAC-A------------------------------CGCACGCGCCCCACA------CAAGCACAC--ACACACACACACATTGA----GTGTATGTGGG----------------------------------TGTGTG--TATAC-A-AGGCATACAAACGCAAGCTGCAGAAGTGCGTTCGA---AAACAGAA |
| droEug1 |
scf7180000407252:136371-136508 - |
|
ACCAATGCAAACAC--------ACCCA------------------------------------------CACAACCACAA------------------------------CGCACGCGCCCCACACAC----AAATA----------C--AAGGAATGTCTGTG--------TGT-------------------GTGCTTGAGTTTGT---GTT--TATAC-AAAAGCATACAAACGCAAGCTGCAAAAGTGCGTTCGA---AAACAGAA |
| droBia1 |
scf7180000302188:160512-160641 - |
|
ACCAATGCAAACAC----------CCA------------------------------------------CACAACCACAA------------------------------CGCACGCGCCCCACACAC--------------AGATAC------AGAGTTTGCG--------GGA-------------------GTGTGTGTGTCG-----CTG--TATAC-AAAGGCATACAAACGCAAGCTGCAAAAGTGCGTTCGA---AAACAGAA |
| droTak1 |
scf7180000415779:57497-57619 + |
|
ACCAATGCAAACAC----------CCA------------------------------------------CACAATCAC-A------------------------------CGCACGCGCCCCACA------C----------ACACAGATACGGTGCGATTGT--------GTG----------------------------------TGTGTG--TATAC-AAAAGCATACAAACGCAAGCTGCAAAAGTGCGTTCGA---AAACAGAA |
| droEle1 |
scf7180000491273:1727164-1727287 - |
|
ACCAATGCAAACAC----------ACA------------------------------------------CACAAGCGCAA------------------------------CGCACGCGCCCCACA------CAAATACAT--AGACACAC----AGTATC------------TG----------------------------------TGTGTA--TATGG-AAAAGCATACAAACGCAAGCTGCAAAAGTGCGTTCAA---AAACAGAA |
| droRho1 |
scf7180000767698:404-525 + |
|
ACCAATGCAAACAC----------ACA-------------------------------C-----ACACACACAACAGCAA------------------------------CGCACGCGCCCCACA------CAAATACA----------------GTATCT----------GTG----------------------------------TGTGTA--TATAG-AAGAGCATACAAACGCAAGCTGCAAAAGTGCGTTCAA---AAACAGAA |
| droFic1 |
scf7180000448848:123-246 + |
|
ACCAATGCAAACACTCGCACTCACGCA------------------------------------------CACAACCGCAA------------------------------CGCACGCGCCCCACA------CAAACACAC--GTACACA------------------------------------------------------------GTGTG--TATAC-AAAAGCATACAAACGCAAGCTGCAAAAGTGCGTTCGA---AAACAGAA |
| droKik1 |
scf7180000302643:882873-882990 - |
|
ACCAATGCAAACAC----------ACA-------------------------------AATACAACACACACATACAT-A------------------------------CGCACGCGCCGCACTCAC------ATACC--------C------------------------------------------------------------C--GTA--TATACTAAAAGCATACAAACGCAAGCTGCAAAAGTGCGTTCGAA--AAACAGAA |
| droBip1 |
scf7180000396572:1957527-1957660 - |
|
ACCAATACAAACAC----------ATA------------------------------------------GA-------------------CGCACAAAAACCAATACAAACGCACGCGCCCCACA------CAAACACCC--GAACATACA---------------------TAT--------ATAGTAAGCCC--------------AGTACA--TATAC---AAATATACAAACGCAAGCTGCAAAAGTGCGTTCGA---AA------ |
| dp5 |
4_group2:543412-543591 - |
|
ACCAATACACA--------------GGCAAAAGTAAGGGCGCGTGTCGCACACAAACG-----------CGCACACAC-CTACACATAGACTCATACATACTCATATTTTCGTACATACCATACATAC-------------------------------ATATG--------TATGTACATACATA----------------------CATACA--TACAT-ACATATATACAAACGCAAGCTGCAAAAGTGCGTTCCCCATAGAGAATG |
| droPer2 |
scaffold_8:3347445-3347612 + |
|
ACCAATACACA--------------GGCAAAAGTAAGGGCGCGTGTCGCACACAAACG-----------CACACACAC-CTACACATAGACTCATACATACTCATATATTCGTACATACCATACATAC--------ATAT--G-----------------------------TA----------------------------------CATACA--TACAT-ACATATATACAAACGCAAGCTGCAAAAGTGCGTTCCCCATAGAGAATG |
| droWil2 |
scf2_1100000004521:3509227-3509329 + |
|
AC----------AC----------ACA------------------------------------------CACAAACAA-A------------------------------CGCACGCGCCCCACACACACATATACGCAC--ACACACACACGCACA--------------------------------------------CATTT-----ATGTAT-----------ATACAAGCGCAAGCTGCAAAAGTGCGTTC------------- |
| droVir3 |
scaffold_12970:10205657-10205743 + |
|
TCCAGT----------------------------------------CGCATACACACA-----------CGCACACAC-GCACACACGCACA---------------------------CCTACA------CACACACAC--AC------ACACATAGCCTGTG--------TGC-------------------GTGTGTGTGCCT-----G---------------------------------------------------------- |
| droMoj3 |
scaffold_6500:27080503-27080633 + |
|
ACACACACACACAC----------ACA------------------------------------------CACACACAC-A------------------------------CACACACAC---A--------CACACACACACACACAC--ACACAATACATGTG--------TGT-------------------ATGTATAT---------GTG--TATAT-AAAGATATATATACATATG-TACATAAGTACGCCCAC---ATACACAG |
| droGri2 |
scaffold_15110:13283685-13283783 + |
|
ACTAACGCACACAC----------ACA------------------------------------------CCAACACAC-ACACACGG-----------------------------AGACACACA------CACACGCAC--ACACACATACTCACTACTTGTG--------TGT-------------------GTGTGTGTGTGT-------------------------------------------GTGTGTGCGA----------- |